Impact of genetic polymorphisms on platelet function and response to anti platelet drugs
In the last decades cardiovascular genomic have gained increasing interest. This consists in the identification of polymorphic genes responsible for the susceptibility to cardiovascular disease including coronary artery disease (CAD) (1,2). The awareness that early identification of these subjects at risk is pivotal to optimize the treatment stems from the evidence that 15% of myocardial infarctions (MI) and coronary revascularizations may be attributed to family history irrespectively from other conventional risk factors; in addition, the risk of premature CAD is three-times higher in first degree relatives than in the general population (3,4). The interest in cardiovascular genomics has grown in parallel with the outbreak of advanced techniques developed to analyse human genetic footprints. Researchers nowadays have shifted from genetic linkage analysis and a candidate gene approach toward genome-wide association (GWAS) studies and the analysis of miRNA-mRNA expression profiles.
Genes involved in platelet activation and aggregation play a key role in the predisposition to CAD. Many evidences support that platelet hyper-reactivity is linked to the risk of MI and moreover platelets volume is an independent predictor of outcomes after an ischemic event (5,6). Studies conducted on healthy volunteers demonstrated the existence of a considerable inter-variability of platelet response to agonists and in particular the hyper-reactivity phenotype seems to be heritable (7). Moreover, the FRE study investigators demonstrated in a sample of twins how heritable factors were responsible for 21–30% of agonist-induced platelet aggregation whereas other common cardiovascular risk factors played a minor role (8).
Polymorphic genes involved in platelet function
Platelets glycoproteins
Multiple glycoproteins on platelets surface are responsible for the complex cascade leading to blood clotting. Several candidate gene association studies focused on these glycoproteins because every mutation in genes encoding for these proteins can be potentially responsible for altered platelet reactivity. Hereby we describe the most known and well-studied polymorphisms responsible for susceptibility to CAD.
GP1BA and ITGA2 genes encode respectively for the glycoprotein Ib and the integrin α2β1 which are responsible for the platelet-binding to collagen. In the first gene three polymorphisms have been demonstrated to increase the risk of CAD: the VNTR (variable number of tandem repeats of a sequence of 13-aminoacids), -5C allele (C/T substitution five nucleotides before the ATG starting codon), and the Met145 (Met/Thr in the residue 145). For the second gene the T-807 allele increases platelet aggregation and accelerates the occurrence of cardiovascular accidents (9,10). GP6 encodes for the glycoprotein VI a receptor for collagen that plays a critical role in collagen-induced platelet aggregation and the dimorphism T13254C has been also associated with the risk of MI (11).
The PLA2 polymorphism (HPA-1) of the gene ITGA2B determines the substitution Pro/Leu in position 33 of the glycoprotein IIIa and is responsible for an increased platelet aggregation, a lower threshold of response to agonists and a weak response to anti-platelets drugs. Important clinical implications have been demonstrated for this polymorphism: higher and premature risk of MI, higher risk of stroke in hypertensive patients and increased incidence of major events in patient with known CAD (12). Furthermore, among glycoproteins influencing platelets adhesion and aggregation there is the GpIb/IX whose single nucleotide polymorphism (SNP) T524C confers higher risk of CAD (13).
CD62P and PSGL-1, the genes encoding for the P-selectin and its ligand PSGL-1, have been extensively studied as candidate genes related to ischemic heart disease. PCR sequencing techniques have recognized 13 SNPs but only the T715P polymorphism is associated with the risk of MI as confirmed by a case control study in which it was demonstrated that the T715 allele is more frequent in CAD patients especially in those with hypercholesterolemia (14).
Thus so far a large number of case-control studies highlighted the association between platelets glycoproteins SNPs and CAD susceptibility. Yet, these associations fail to be confirmed when reassessed on a larger sample sized meta-analysis, therefore raising doubts on the robustness of these evidences (15).
Platelets receptors
Many agonists and antagonists are able to enhance or decrease platelets aggregation by binding surface receptors encoded by highly polymorphic genes.
Adrenergic receptors alpha and beta are both expressed on platelets surface. Even if epinephrine is a weak agonist, the variant 6.3 kb of the α2A receptor determines an increased response and higher platelet reactivity in patients with CAD treated with dual antiplatelet therapy (DAPT) (16). The genetic variant of ADRA2B gene responsible for the deletion of three glutamate in the α2B receptor gives resistance to down-regulation, resulting in a higher risk of acute coronary events. Likewise also for the ADRB2 have been described genetic variants that influence β receptor expression with important clinical implications: e.g., the alleles-G79 and -A491 have been associated with the presence and severity of CAD (17-19). Nitric oxide synthase 3 plays a key role in platelets function by reducing aggregation. The polymorphisms Glu298Asp and intron4 (an allele) contribute to atherosclerosis development and risk of CAD and also influence platelet reactivity in patients with acute coronary syndromes (ACS) on DAPT (20,21). Furthermore it was recently demonstrated that the effect of Glu298Asp is offset by a loading dose of DAPT in stable CAD patients undergoing elective PCI (22).
Protease-activated receptors 1 and 4 (PAR-1, -4) are mainly involved in thrombin-mediated platelets activation and their polymorphisms are associated with increased platelet aggregation. In particular, the dimorphism A/T in the intervening sequence 14 determines increased PAR-1 expression and the A-allele homozygosity causes platelets hyper-reactivity despite clopidogrel in CAD patients (23). Moreover the dimorphism G/A in locus 1738 is another important polymorphism that was demonstrated to increase the risk of MI.
Prostaglandin H2 in platelets is metabolized in thromboxane (TXA2) by thromboxane synthase and the binding to its receptor TXA2R increases platelet aggregation. Interestingly, in a case-control study its polymorphism rs768963 was demonstrated to be more frequent in patients with cerebral infarction while the polymorphism rs1131882 was associated with a higher risk of carotid plaque vulnerability (24).
Directly linked to the thromboxane pathway there is the platelet endothelial aggregation receptor 1 (25). Genome wide associated studies first found a strong association between a specific region on the chromosome 1q23 and cardiovascular events and thereafter further analyses revealed the presence in this region of the polymorphism rs12041331 of PEAR1. This polymorphism is able to influence response to antiplatelet drugs and A-allele carriers have an increased risk of MI. More recently other PEAR1 genetic variations were associated with response to aspirin and clopidogrel in CAD patients: rs11264580, rs2644592, rs3737224 (26).
Lastly, C2238 polymorphism of the gene encoding for the atrial natriuretic peptide (ANP) has been associated to higher risk of cerebrovascular accidents and MI. An increased platelet aggregation has been proposed among the possible mechanisms underlying the untoward cardiovascular outcomes in carriers of the C2238 genetic variant (27).
Newly discovered genes involved in platelet function
With the advent of GWAS it has been possible to find the inherited basis of CAD from a wider perspective without a pre-defined hypothesis as the case of candidate gene studies. This approach consists in the analysis of thousands of genomes with the aim to detect variants that occur at different frequencies in cases and controls. Since the first GWAS discovery in 2007 that identified a locus on chromosome 9p21, so far 32 loci have been associated with the risk of CAD and MI. Interestingly these loci are related to genes whose role is frequently unknown in the pathogenesis of the disease or that in many cases are embedded in non-coding regions (28), suggesting that these variants are more likely to affect gene regulation rather than protein structure. Exome-wide association studies are the new trend and, differently from GWAS, they analyse only variants occurring in the coding regions (29). This approach revealed the involvement of the genes affected by risk alleles in different pathophysiological pathways, including also platelets aggregation. For instance recently an exome-sequencing study demonstrated a digenic mutation in GUCYA3 (rs7692387 chromosome 4) a subunit of the soluble form of guanylyl cyclase, which is the major receptor of NO. This subunit is responsible for accelerated thrombus formation and is associated with high prevalence of premature CAD and MI (30). Furthermore, another variant associated with CAD is rs3184504 (chromosome 12) tagging the SH2B3 which encodes for an adaptor protein expressed also in leucocytes and platelets. The risk allele at the SH2B3 locus is associated with increased platelet count and moreover in a knock-out mouse model increased platelet counts and reduced thrombus stability were found (31). Accordingly to the idea of focusing the attention on polymorphisms in the coding regions, recently a study of genome-wide platelet RNA expression was conducted and identified the association between polymorphisms of COMMD7 and LRRFIP1 and a modest susceptibility to MI. These genes encode for proteins regulating platelets gene expression (32).
Lastly, the microRNAs (33) deserve particular attention, as much like DNA polymorphisms, they can be considered biomarker of platelets function.
miRNAs are 22-nucleotides non-coding RNAs able to regulate processing and transcription of mRNA and thus gene expression in platelets that are anucleated cells. Different expression of miRNAs can account also for racial variability in platelets activation, indeed it was found that miR376-c regulates platelets activation PAR-4 mediated, which is responsible of the higher responsiveness of platelets in Blacks (34).
miRNAs can be stored in platelets or may be transported in microparticles that originating from platelets surface are then released into bloodstream, thus may reach different cells type influencing their gene expression. This is the case of miR320-b that can inhibit the expression of ICAM-1 in cultured endothelial cells. More interestingly a recent study conducted in STEMI patients found that miR-22, miR-185, miR-320-b, miR-425-5p were reduced in platelets in comparison with controls and their content in the thrombus was even lower, suggesting that during MI these miRNAs are released from platelets and may influence thrombus formation (35).
Circulating platelets miRNAs have been also associated with diabetes, MI risk and smoking. In particular plasma miRNAs levels seem to be lower in healthy subjects taking anti-platelets medications. Lastly miR-223 is one of the most studied because many evidences demonstrated its association with platelets function and CAD. For instance circulating levels of miR-223 are lower in patients with troponine negative NSTEMI and furthermore were lower in CAD patients with high residual platelet reactivity on treatment with clopidogrel (36-38).
Polymorphic genes involved in response to anti-platelets drugs
Pharmacogenetics is the branch of pharmacology involved in the study of genetic factors influencing the response to drugs. As for many other cardiovascular drugs also for anti-platelets there is a great variability of response between individuals and at least part of this can be attributed to highly polymorphic genes involved in drugs metabolism. An optimal inhibition of platelet aggregation is crucial in CAD patients but especially in those undergoing PCI and stent implantation, indeed the evidence that patients with high on treatment platelet reactivity experience worse outcomes, related to ischemic events and stent thrombosis, triggered the interest in the identification of genetic determinants of the so called anti-platelets resistance.
Aspirin resistance
Prevalence of aspirin resistance varies widely depending on the dosage and on the method used for its assessment. In studies using 100 mg or less there was a higher prevalence in comparison with those using 300 mg or more (36% vs. 19%) (39). Likewise, when using point of care devices to analyse platelets function the resistance was higher than that measured with the light transmittance aggregometry (26% vs. 6%). Clinical outcomes related to aspirin resistance in CAD patients have been extensively studied, and besides the assessment method or the definition used, all demonstrated an increase of ischemic events in patients who underwent to elective PCI. Nonetheless high loading dose of aspirin before PCI seems to overcome the variability in platelet response (40). Even if the relation is still controversial, the putative polymorphisms responsible for aspirin resistance are COX1 (A842G, C50T), P1A1/A2, GPIa (C807T), P2Y12 (H1/H2), P2Y1 (8A1622G) and moreover in diabetic patients ADRA2A (rs4311994), TXBA2R (rs1131882), PLA2G7 (rs7756935), 9p21.3 (rs10120688) (41,42).
Clopidogrel resistance
High platelet reactivity despite treatment with clopidogrel may be attributed to genetic polymorphisms of the P2Y12 receptor or of enzymes involved in its metabolism such as CYP, ABC and PON (Table 1).
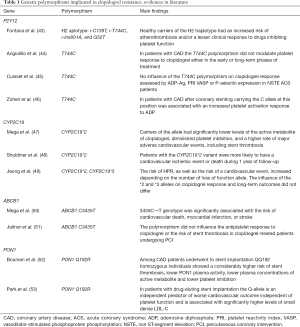
Full table
P2Y12 is the ADP receptor expressed on platelets surface and when clopidogrel irreversibly binds to it makes impossible the bond of its natural ligand thus inhibiting platelets aggregation. This receptor is highly polymorphic and five SNPs have been found to increase the risk of CAD: C139T, T744C, an insertion in position 801, C34T and G52T transversion. Because these genetic variants were found in linkage disequilibrium two haplotypes can be defined: H1 (139C, 744T, 801-A, 52G) and H2 (139T, 744C, 801A, 52T). The latter haplotype has been associated with higher platelet aggregation and the presence of PAD. Nonetheless, evidences in CAD patients are still controversial indeed it was found a lack of association between higher platelets aggregation and for instance T744C in both stable CAD and NSTEMI patients (44,45) whereas more recently another study demonstrated higher prevalence of this allele in patients with ACS than in controls (46).
At the opposite the association between clopidogrel resistance and polymorphisms of CYP enzymes is ascertained. In particular, CYP2C19 is the major enzyme responsible of clopidogrel transformation in its active metabolite. Even if up to 25 SNPs have been identified for this gene, the CYP2C19*2 and the CYP2C19*3 are the most studied allelic variants responsible for the loss of function of the enzyme, while the CYP2C19*17 allele seems to be associated with a gain of function. Nonetheless, recent findings suggest a role also for the *4 and *8 alleles in clopidogrel response variability (54).
The *2 allele frequencies are 15% in Caucasians and Africans and 29–35% in Asians (54). The *2 and *17 alleles are in linkage disequilibrium and based on the combination of these two, patients can be defined as ultra-rapid, extensive, intermediate or poor clopidogrel metabolizer (55,56). Patients with at least one loss of function allele have a decrease of 32% of the active metabolite (47) and moreover their association with clopidogrel response seems to be gene-dose dependent (48). These evidences led the FDA to publish a boxed warning on clopidogrel label for poor and intermediate metabolizers and in these patients antiplatelets other than clopidogrel are recommended (54). Since the GRAVITAS study demonstrated that in patients with high on treatment platelet reactivity undergoing PCI an increase of clopidogrel dosage (600 mg loading dose +150 mg daily) doesn’t reduce the endpoint of CV death, non-fatal MI and stent thrombosis, therefore the only useful strategy in these patients is the treatment with alternative P2Y12 inhibitors such as prasugrel or ticagrelor (57). Many studies highlighted the influence of the CYP2C19 genotype on platelet reactivity in CAD patients. In particular, the *2 allele is responsible of high on treatment platelet reactivity in patients taking DAPT after PCI and stenting while response to aspirin is not influenced (58). Furthermore, the *3 allele also plays a pivotal role and its influence on clopidogrel resistance is greater than that of the *2 allele in Asians (49). Consistently, both the TRITON-TIMI 38 and the PLATO studies demonstrated that carriers of the *2 allele treated with clopidogrel incurred in more cardiovascular events than non-carriers (42). Nonetheless studies including patients at lower risk, with lower rates of PCI and stenting or treated with clopidogrel for atrial fibrillation and stroke failed to demonstrate the association between the *2 allele and CV events (54). For this reason the Clinical Pharmacogenetics Implementation Consortium recommends a CYP2C19-genotype guided therapy only in patients with ACS or undergoing to PCI and not in all patients treated with clopidogrel, even if the decision to perform or not CYP2C19 screening before starting the therapy with clopidogrel is left up to the individual clinician (54). The usefulness in identifying the clopidogrel metabolic profile (slow and rapid metabolizers) has been recently rejected by the ARCTIC-GENE study, where slow metabolizer didn’t experience more composite death, MI, stent thrombosis, stroke or urgent revascularization one year after stent implantation. The genotyping to identify loss of function alleles well predicted the platelets response to clopidogrel although was not related to clinical outcomes (59). Moreover, the monitoring of platelets function and the adjustment of treatment as well as in the ARCTIC trial didn’t improve outcomes (60). Interestingly in 32% of rapid metabolizers of the ARCTIC-GENE study was found a poor response to clopidogrel supporting the hypothesis that the CYP2C19 genetic profile is not the only factor involved in high on treatment platelet reactivity. Drugs interaction plays an important role in clopidogrel resistance, especially the interaction with proton pump inhibitor (61). The CYP2C19, CYP3A4, CYP2C9 are the enzymes involved in their metabolism but also are inhibited by these drugs, interfering in this way with clopidogrel activity. The highest interaction with CYPC2C19 has been demonstrated for esomeprazole and omeprazole while pantoprazole doesn’t seem to attenuate clopidogrel response (62), therefore the interaction with clopidogrel cannot be considered a class effect (61). Strategies such as increasing clopidogrel dosages or staggered intake were unsuccessful in overcoming the interaction omeprazole-clopidogrel (63,64). Moreover many studies tried to address the issue if this interaction may influence clinical outcomes. The COGENT study found that the association omeprazole and clopidogrel led to a reduction in gastrointestinal bleeding but no increased risk of MACE or MI with PPI, nonetheless the study was stopped earlier for lack of funding, so the days of follow-up were less than expected and also the number of MACE was low (65). Even if in TRITON-TIMI 38 trial, no association existed between use of PPI and cardiovascular death, MI or stroke in clopidogrel-treated patients, subsequent meta-analysis showed that the combination of clopidogrel and PPI increased the risk of MACE and stent thrombosis and MI after PCI, with no impact on long-term mortality and all cause mortality (66,67).
ABCB1 gene encodes for the P-glycoprotein expressed in intestinal epithelial cells and responsible for the transition of clopidogrel from the intestinal lumen into the bile, thus limiting clopidogrel absorption and bioavailability. In the pharmacogenetic analysis of the TRITON-TIMI 38 the polymorphism C3435T and in particular the T allele was demonstrated to be an independent predictor of primary endpoint (cardiovascular death, MI or stroke) (50). Nonetheless with regard to this association there are still doubts raised by other discordant studies (51,68,69).
PON1 is the gene encoding for the paraoxonase 1, a calcium-dependent anti-oxidant enzyme released by the liver into the circulation and carried by high-density lipoproteins that is involved in the bioactivation of clopidogrel. The Q192R polymorphism is able to influence clopidogrel response, indeed the QQ192 homozygous patients have lower PON activity, lower activated clopidogrel, higher platelet reactivity and higher risk of stent thrombosis (52). Also in this case results are not univocal indeed other studies confirmed the association of this polymorphism with worse outcome after PCI (53) while others failed to demonstrate the same association between PON polymorphism and platelet reactivity (70,71).
Prasugrel and ticagrelor
Like clopidogrel, prasugrel is a pro-drug and its bio-activation requires intestinal esterase and enzymes CYP3A4 and CYP2B6 and to a lesser degree CYP2C19 and CYP2C9. Several studies investigated the role of CYP polymorphisms on prasugrel activity although results are inconclusive (72-76). Furthermore in the TRITON-TIMI 38 trial the relation between clinical outcomes and prasugrel or clopidogrel was analysed in both carriers and non-carriers of the loss of function allele of CYP2C19 and results showed that prasugrel in comparison with clopidogrel reduced the risk of ischemic events of 19% in carriers patients, whereas the occurrence of ischemic events was similar in non-carriers (75). Nevertheless, besides the influence of such polymorphisms, the prevalence of high on treatment platelet reactivity (HTPR) in patients treated with prasugrel after an ACS is in the range of 4–6% and the major determinants of this are obesity and multivessel disease (77). Possible strategies to overcome prasugrel low responsiveness are to increase the maintenance dose to 20 mg/die or switching to ticagrelor (73,78).
Ticagrelor is a non-thienopyridine reversible antagonist of P2Y12 and because it doesn’t need bio-activation, it has a rapid onset of action. Consistent data regarding the influence of P2Y12 genetic polymorphisms demonstrated that ticagrelor response is not affected by the polymorphism rs5443 of the G-protein polypeptide 3 (79,80). Ticagrelor is more effective than clopidogrel in reducing primary endpoints as demonstrated in the PLATO study, moreover ticagrelor overcomes clopidogrel resistance enabling to reach platelet reactivity values well below those associated with ischemic events in both clopidogrel responders and non-responders (81).
These evidences suggest that in order to avoid antiplatelets low responsiveness prasugrel or ticagrelor should be the first choice although it should be kept in mind that the greater effectiveness is reached at the cost of a higher risk of bleedings as reported in the TRITON-TIMI38 and in the PLATO studies (82).
In the recent PHARMCLO trial ACS patients were randomized to receive the antiplatelet therapy (clopidogrel, prasugrel or ticagrelor) based on patients’ characteristics (standard of care arm) or based on testing of ABCB1, CYP2C19*2, and CYP2C19*17(pharmacogenomics arm). Even if the study was prematurely stopped due to the lack of in vitro diagnosis certification for the genotyping instrument and was underpowered for the stent thrombosis, the results showed that the pharmacogenomics arm after 1-year follow-up experienced less ischemic and bleeding events (83).
New perspectives
While investigators seem to have abandoned the strategy of platelet reactivity assessment to guide antiplatelet therapy, so far with the advent of new technologies enabling the genotyping easily, without any particular skill and trough point of care tests, it seems to be wiser to avoid a useless and potentially unsafe overshooting with more potent drugs as prasugrel and ticagrelor and to genotype the patient in order to prescribe the most suitable antiplatelet therapy for him. A waiting time of about one hour for the result of the genotype test could prevent us to face in our patients dangerous bleedings. Elderly and frail patients are the ones who can most benefit of this approach, indeed while there is a trend to empirically treat them less aggressively by choosing a less potent antiplatelet therapy nonetheless the fear of an insufficient platelet inhibition and of ischemic events is always present.
Another aspect that should be considered is the pharmacoeconomic one, indeed prasugrel and ticagrelor are not only more potent drugs but also more expensive, so their extensive prescription to anyone who needs a dual antiplatelet therapy seems to be a waste of resources. Therefore a genotype- based approach is useful to allocate prasugrel and ticagrelor only to those who can most benefit from them. Nonetheless also the genotyping is not cost-free so now the results of the POPular Genetics study are awaited with great interest. This is an on-going large-scale trial analysing the cost-effectiveness, the net clinical benefit and the safety of a CYP2C19 genotype-guided antiplatelet therapy compared to a non-tailored strategy in STEMI patients (84). So far as this study will demonstrate that prescribing the cheapest anti-P2Y12 clopidogrel in absence of a genetic resistance is safe and effective as prescribing to all prasugrel or ticagrelor and furthermore that the money saved in this way overweight the cost of genotyping, this will lead toward a shift from trying to discover ever more potent antiplatelet drugs and toward choosing the right therapy for each individual patient.
Acknowledgements
T Strisciuglio and C De Biase are supported by the Cardiopath PhD programme.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Wung SF, Hickey KT, Taylor JY, et al. Cardiovascular genomics. J Nurs Scholarsh 2013;45:60-8. [Crossref] [PubMed]
- Gigante B, Bellis A, Visconti R, et al. Retrospective analysis of coagulation factor II receptor (F2R) sequence variation and coronary heart disease in hypertensive patients. Arterioscler Thromb Vasc Biol 2007;27:1213-9. [Crossref] [PubMed]
- Andresdottir MB, Sigurdsson G, Sigvaldason H, et al. Fifteen percent of myocardial infarctions and coronary revascularizations explained by family history unrelated to conventional risk factors. The Reykjavik Cohort Study. Eur Heart J 2002;23:1655-63. [Crossref] [PubMed]
- Vaidya D, Yanek LR, Moy TF, et al. Incidence of coronary artery disease in siblings of patients with premature coronary artery disease: 10 years of follow-up. Am J Cardiol 2007;100:1410-5. [Crossref] [PubMed]
- Trip MD, Cats VM, van Capelle FJ, et al. Platelet hyperreactivity and prognosis in survivors of myocardial infarction. N Engl J Med 1990;322:1549-54. [Crossref] [PubMed]
- Boos CJ, Lip GY. Assessment of mean platelet volume in coronary artery disease - what does it mean? Thromb Res 2007;120:11-3. [Crossref] [PubMed]
- Yee DL, Sun CW, Bergeron AL, et al. Aggregometry detects platelet hyperreactivity in healthy individuals. Blood 2005;106:2723-9. [Crossref] [PubMed]
- Gaxiola B, Friedl W, Propping P. Epinephrine induced platelet aggregation. A twin study. Clin Genet 1984;26:543-8. [Crossref] [PubMed]
- Esen FI, Hancer VS, Küçükkaya RD, et al. Glycoprotein Ib-alpha Kozak polymorphism in ischemic stroke. Neurol Res 2012;34:68-71. [Crossref] [PubMed]
- Kunicki TJ. The influence of platelet collagen receptor polymorphisms in hemostasis and thrombotic disease. Arterioscler Thromb Vasc Biol 2002;22:14-20. [Crossref] [PubMed]
- Ollikainen E, Mikkelsson J, Perola M, et al. Platelet membrane collagen receptor glycoprotein VI polymorphism is associated with coronary thrombosis and fatal myocardial infarction in middle-aged men. Atherosclerosis 2004;176:95-9. [Crossref] [PubMed]
- Galasso G, Santulli G, Piscione F, et al. The GPIIIA PlA2 polymorphism is associated with an increased risk of cardiovascular adverse events. BMC Cardiovasc Disord 2010;10:41. [Crossref] [PubMed]
- Abboud N, Amin H, Ghazouani L, et al. Polymorphisms of human platelet alloantigens HPA-1 and HPA-2 associated with severe coronary artery disease. Cardiovasc Pathol 2010;19:302-7. [Crossref] [PubMed]
- Bugert P, Vosberg M, Entelmann M, et al. Polymorphisms in the P-selectin (CD62P) and P-selectin glycoprotein ligand-1 (PSGL-1) genes and coronary heart disease. Clin Chem Lab Med 2004;42:997-1004. [Crossref] [PubMed]
- Ni W, He J, Wang H, et al. Association of Platelet Membrane Glycoprotein HPA-2a/b, GP VI T13254C, and GP Ibα VNTR Polymorphisms with Risk of Coronary Artery Disease: A Meta-Analysis. Biomed Res Int 2017;2017. [Crossref] [PubMed]
- Peace AJ, Mangiacapra F, Bailleul E, et al. α2A-Adrenergic receptor polymorphism potentiates platelet reactivity in patients with stable coronary artery disease carrying the cytochrome P450 2C19*2 genetic variant. Arterioscler Thromb Vasc Biol 2014;34:1314-9. [Crossref] [PubMed]
- Barbato E, Berger A, Delrue L, et al. GLU-27 variant of beta2-adrenergic receptor polymorphisms is an independent risk factor for coronary atherosclerotic disease. Atherosclerosis 2007;194:e80-6. [Crossref] [PubMed]
- Piscione F, Iaccarino G, Galasso G, et al. Effects of Ile164 polymorphism of beta2-adrenergic receptor gene on coronary artery disease. J Am Coll Cardiol 2008;52:1381-8. [Crossref] [PubMed]
- Yamada Y, Izawa H, Ichihara S, et al. Prediction of the risk of myocardial infarction from polymorphisms in candidate genes. N Engl J Med 2002;347:1916-23. [Crossref] [PubMed]
- Fatini C, Sticchi E, Bolli P, et al. eNOS gene influences platelet phenotype in acute coronary syndrome patients on dual antiplatelet treatment. Platelets 2009;20:548-54. [Crossref] [PubMed]
- Strisciuglio T, Di Gioia G, De Biase C, et al. Genetically Determined Platelet Reactivity and Related Clinical Implications. High Blood Press Cardiovasc Prev 2015;22:257-64. [Crossref] [PubMed]
- Strisciuglio T, Barbato E, De Biase C, et al. T2238C Atrial Natriuretic Peptide Gene Variant and the Response to Antiplatelet Therapy in Stable Ischemic Heart Disease Patients. J Cardiovasc Transl Res 2018;11:36-41. [Crossref] [PubMed]
- Dupont A, Fontana P, Bachelot-Loza C, et al. An intronic polymorphism in the PAR-1 gene is associated with platelet receptor density and the response to SFLLRN. Blood 2003;101:1833-40. [Crossref] [PubMed]
- Yi X, Lin J, Luo H, et al. Genetic variants of PTGS2, TXA2R and TXAS1 are associated with carotid plaque vulnerability, platelet activation and TXA2 levels in ischemic stroke patients. PLoS One 2017;12. [Crossref] [PubMed]
- Lewis JP, Ryan K, O'Connell JR, et al. Genetic variation in PEAR1 is associated with platelet aggregation and cardiovascular outcomes. Circ Cardiovasc Genet 2013;6:184-92. [Crossref] [PubMed]
- Yao Y, Tang XF, Zhang JH, et al. Association of PEAR1 genetic variants with platelet reactivity in response to dual antiplatelet therapy with aspirin and clopidogrel in the Chinese patient population after percutaneous coronary intervention. Thromb Res 2016;141:28-34. [Crossref] [PubMed]
- Carnevale R, Pignatelli P, Frati G, et al. C2238 ANP gene variant promotes increased platelet aggregation through the activation of Nox2 and the reduction of cAMP. Sci Rep 2017;7:3797. [Crossref] [PubMed]
- Maouche S, Schunkert H. Strategies beyond genome-wide association studies for atherosclerosis. Arterioscler Thromb Vasc Biol 2012;32:170-81. [Crossref] [PubMed]
- Kessler T, Vilne B, Schunkert H. The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease. EMBO Mol Med 2016;8:688-701. [Crossref] [PubMed]
- Erdmann J, Stark K, Esslinger UB, et al. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature 2013;504:432-6. [Crossref] [PubMed]
- Takizawa H, Nishimura S, Takayama N, et al. Lnk regulates integrin alphaIIbbeta3 outside-in signaling in mouse platelets, leading to stabilization of thrombus development in vivo. J Clin Invest 2010;120:179-90. [Crossref] [PubMed]
- Goodall AH, Burns P, Salles I, et al. Transcription profiling in human platelets reveals LRRFIP1 as a novel protein regulating platelet function. Blood 2010;116:4646-56. [Crossref] [PubMed]
- Nagalla S, Shaw C, Kong X, et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood 2011;117:5189-97. [Crossref] [PubMed]
- Edelstein LC, Simon LM, Montoya RT, et al. Racial differences in human platelet PAR4 reactivity reflect expression of PCTP and miR-376c. Nat Med 2013;19:1609-16. [Crossref] [PubMed]
- Gidlöf O, van der Brug M, Ohman J, et al. Platelets activated during myocardial infarction release functional miRNA which can be taken up by endothelial cells and regulate ICAM1 expression. Blood 2013;121:3908-17, S1-26.
- Badrnya S, Baumgartner R, Assinger A. Smoking alters circulating plasma microvesicle pattern and microRNA signatures. Thromb Haemost 2014;112:128-36. [Crossref] [PubMed]
- Zhang YY, Zhou X, Ji WJ, et al. Decreased circulating microRNA-223 level predicts high on-treatment platelet reactivity in patients with troponin-negative non-ST elevation acute coronary syndrome. J Thromb Thrombolysis 2014;38:65-72. [Crossref] [PubMed]
- Chyrchel B, Totoń-Żurańska J, Kruszelnicka O, et al. Association of plasma miR-223 and platelet reactivity in patients with coronary artery disease on dual antiplatelet therapy: A preliminary report. Platelets 2015;26:593-7. [Crossref] [PubMed]
- Hovens MM, Snoep JD, Eikenboom JC, et al. Prevalence of persistent platelet reactivity despite use of aspirin: a systematic review. Am Heart J 2007;153:175-81. [Crossref] [PubMed]
- Mangiacapra F, Barbato E. Individual variability of response to antiplatelet therapy is an important determinant of adverse clinical outcome. High Blood Press Cardiovasc Prev 2010;17:121-30. [Crossref]
- Postula M, Kaplon-Cieslicka A, Rosiak M, et al. Genetic determinants of platelet reactivity during acetylsalicylic acid therapy in diabetic patients: evaluation of 27 polymorphisms within candidate genes. J Thromb Haemost 2011;9:2291-301. [Crossref] [PubMed]
- Tantry US, Jeong YH, Navarese EP, et al. Influence of genetic polymorphisms on platelet function, response to antiplatelet drugs and clinical outcomes in patients with coronary artery disease. Expert Rev Cardiovasc Ther 2013;11:447-62. [Crossref] [PubMed]
- Fontana P, Dupont A, Gandrille S, et al. Adenosine diphosphate–induced platelet aggregation is associated with P2Y12 gene sequence variations in healthy subjects. Circulation 2003;108:989-95. [Crossref] [PubMed]
- Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, et al. Lack of association between the P2Y12 receptor gene polymorphism and platelet response to clopidogrel in patients with coronary artery disease. Thromb Res 2005;116:491-7. [Crossref] [PubMed]
- Cuisset T, Frere C, Quilici J, et al. Role of the T744C polymorphism of the P2Y12 gene on platelet response to a 600-mg loading dose of clopidogrel in 597 patients with non-ST-segment elevation acute coronary syndrome. Thromb Res 2007;120:893-9. [Crossref] [PubMed]
- Zoheir N, Abd Elhamid S, Abulata N, et al. P2Y12 receptor gene polymorphism and antiplatelet effect of clopidogrel in patients with coronary artery disease after coronary stenting. Blood Coagul Fibrinolysis 2013;24:525-31. [Crossref] [PubMed]
- Mega JL, Close SL, Wiviott SD, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med 2009;360:354-62. [Crossref] [PubMed]
- Shuldiner AR, O'Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 2009;302:849-57. [Crossref] [PubMed]
- Jeong YH, Tantry US, Kim IS, et al. Effect of CYP2C19*2 and *3 loss-of-function alleles on platelet reactivity and adverse clinical events in East Asian acute myocardial infarction survivors treated with clopidogrel and aspirin. Circ Cardiovasc Interv 2011;4:585-94. [Crossref] [PubMed]
- Mega JL, Close SL, Wiviott SD, et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial: a pharmacogenetic analysis. Lancet 2010;376:1312-9. [Crossref] [PubMed]
- Jaitner J, Morath T, Byrne RA, et al. No association of ABCB1 C3435T genotype with clopidogrel response or risk of stent thrombosis in patients undergoing coronary stenting. Circ Cardiovasc Interv 2012;5:82-8, S1-2.
- Bouman HJ, Schömig E, van Werkum JW, et al. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat Med 2011;17:110-6. [Crossref] [PubMed]
- Park KW, Park JJ, Kang J, et al. Paraoxonase 1 gene polymorphism does not affect clopidogrel response variability but is associated with clinical outcome after PCI. PLoS One 2013;8. [Crossref] [PubMed]
- Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 2013;94:317-23. [Crossref] [PubMed]
- Price MJ, Murray SS, Angiolillo DJ, et al. Primary results from genotype information and functional testing a prospective pharmacogenomic analysis of clopidogrel therapy. Presented at: American College of Cardiology Meeting. New Orleans, LA, USA, 2011.
- Beitelshees AL, Horenstein RB, Vesely MR, et al. Pharmacogenetics and clopidogrel response in patients undergoing percutaneous coronary interventions. Clin Pharmacol Ther 2011;89:455-9. [Crossref] [PubMed]
- Price MJ, Berger PB, Teirstein PS, et al. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: the GRAVITAS randomized trial. JAMA 2011;305:1097-105. [Crossref] [PubMed]
- Gurbel PA, Shuldiner AR, Bliden KP, et al. The relation between CYP2C19 genotype and phenotype in stented patients on maintenance dual antiplatelet therapy. Am Heart J 2011;161:598-604. [Crossref] [PubMed]
- Collet JP, Hulot JS, Cuisset T, et al. Genetic and platelet function testing of antiplatelet therapy for percutaneous coronary intervention: the ARCTIC-GENE study. Eur J Clin Pharmacol 2015;71:1315-24. [Crossref] [PubMed]
- Collet JP, Cuisset T, Rangé G, et al. Bedside monitoring to adjust antiplatelet therapy for coronary stenting. N Engl J Med 2012;367:2100-9. [Crossref] [PubMed]
- Fernando H, Dart AM, Peter K, et al. Proton pump inhibitors, genetic polymorphisms and response to clopidogrel therapy. Thromb Haemost 2011;105:933-44. [Crossref] [PubMed]
- Simon N, Finzi J, Cayla G, et al. Omeprazole, pantoprazole, and CYP2C19 effects on clopidogrel pharmacokinetic-pharmacodynamic relationships in stable coronary artery disease patients. Eur J Clin Pharmacol 2015;71:1059-66. [Crossref] [PubMed]
- Cuisset T, Frere C, Quilici J, et al. Comparison of omeprazole and pantoprazole influence on a high 150-mg clopidogrel maintenance dose the PACA (Proton Pump Inhibitors And Clopidogrel Association) prospective randomized study. J Am Coll Cardiol 2009;54:1149-53. [Crossref] [PubMed]
- Ferreiro JL, Ueno M, Capodanno D, et al. Pharmacodynamic effects of concomitant versus staggered clopidogrel and omeprazole intake: results of a prospective randomized crossover study. Circ Cardiovasc Interv 2010;3:436-41. [Crossref] [PubMed]
- Bhatt DL, Cryer BL, Contant CF, et al. Clopidogrel with or without omeprazole in coronary artery disease. N Engl J Med 2010;363:1909-17. [Crossref] [PubMed]
- Siller-Matula JM, Jilma B, Schrör K, et al. Effect of proton pump inhibitors on clinical outcome in patients treated with clopidogrel: a systematic review and meta-analysis. J Thromb Haemost 2010;8:2624-41. [Crossref] [PubMed]
- Bundhun PK, Teeluck AR, Bhurtu A, et al. Is the concomitant use of clopidogrel and Proton Pump Inhibitors still associated with increased adverse cardiovascular outcomes following coronary angioplasty?: a systematic review and meta-analysis of recently published studies (2012 - 2016). BMC Cardiovasc Disord 2017;17:3. [Crossref] [PubMed]
- Wallentin L, James S, Storey RF, et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet 2010;376:1320-8. [Crossref] [PubMed]
- Su J, Yu Q, Zhu H, et al. The risk of clopidogrel resistance is associated with ABCB1 polymorphisms but not promoter methylation in a Chinese Han population. PLoS One 2017;12. [Crossref] [PubMed]
- Lewis JP, Shuldiner AR. Paraoxonase 1 Q192R variant and clopidogrel efficacy: fact or fiction? Circ Cardiovasc Genet 2012;5:153-5. [Crossref] [PubMed]
- Zhang HZ, Kim MH, Guo LZ, et al. CYP2C19 but not CYP2B6, CYP3A4, CYP3A5, ABCB1, PON1 or P2Y12 genetic polymorphism impacts antiplatelet response after clopidogrel in Koreans. Blood Coagul Fibrinolysis 2017;28:56-61. [Crossref] [PubMed]
- Grosdidier C, Quilici J, Loosveld M, et al. Effect of CYP2C19*2 and *17 genetic variants on platelet response to clopidogrel and prasugrel maintenance dose and relation to bleeding complications. Am J Cardiol 2013;111:985-90. [Crossref] [PubMed]
- Franken CC, Kaiser AF, Krüger JC, et al. Cytochrome P450 2B6 and 2C9 genotype polymorphism--a possible cause of prasugrel low responsiveness. Thromb Haemost 2013;110:131-40. [Crossref] [PubMed]
- Brandt JT, Close SL, Iturria SJ, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost 2007;5:2429-36. [Crossref] [PubMed]
- Mega JL, Close SL, Wiviott SD, et al. Cytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation 2009;119:2553-60. [Crossref] [PubMed]
- Varenhorst C, James S, Erlinge D, et al. Genetic variation of CYP2C19 affects both pharmacokinetic and pharmacodynamic responses to clopidogrel but not prasugrel in aspirin-treated patients with coronary artery disease. Eur Heart J 2009;30:1744-52. [Crossref] [PubMed]
- Cayla G, Cuisset T, Silvain J, et al. Prasugrel monitoring and bleeding in real world patients. Am J Cardiol 2013;111:38-44. [Crossref] [PubMed]
- Bassez C, Deharo P, Pankert M, et al. Effectiveness of switching 'low responders' to prasugrel to ticagrelor after acute coronary syndrome. Int J Cardiol 2014;176:1184-5. [Crossref] [PubMed]
- Li MP, Tang J, Wen ZP, et al. Influence of P2Y12 polymorphisms on platelet activity but not ex-vivo antiplatelet effect of ticagrelor in healthy Chinese male subjects. Blood Coagul Fibrinolysis 2015;26:874-81. [Crossref] [PubMed]
- Storey RF, Melissa Thornton S, Lawrance R, et al. Ticagrelor yields consistent dose-dependent inhibition of ADP-induced platelet aggregation in patients with atherosclerotic disease regardless of genotypic variations in P2RY12, P2RY1, and ITGB3. Platelets 2009;20:341-8. [Crossref] [PubMed]
- Gurbel PA, Bliden KP, Butler K, et al. Response to ticagrelor in clopidogrel nonresponders and responders and effect of switching therapies: the RESPOND study. Circulation 2010;121:1188-99. [Crossref] [PubMed]
- Tapp L, Shantsila E, Lip GY. Role of ticagrelor in clopidogrel nonresponders: resistance is futile? Circulation 2010;121:1169-71. [Crossref] [PubMed]
- Notarangelo FM, Maglietta G, Bevilacqua P, et al. Pharmacogenomic Approach to Selecting Antiplatelet Therapy in Patients With Acute Coronary Syndromes: The PHARMCLO Trial. J Am Coll Cardiol 2018;71:1869-77. [Crossref] [PubMed]
- Bergmeijer TO, Janssen PW, Schipper JC, et al. CYP2C19 genotype-guided antiplatelet therapy in ST-segment elevation myocardial infarction patients-Rationale and design of the Patient Outcome after primary PCI (POPular) Genetics study. Am Heart J 2014;168:16-22.e1. [Crossref] [PubMed]


