Artificial intelligence in pediatric and adult congenital cardiac MRI: an unmet clinical need
Introduction
Cardiac MRI (CMR) has become increasingly important for managing pediatric and adult congenital heart disease (ACHD). Its non-invasive nature, lack of ionizing radiation, and 3D anatomic coverage makes it the preferred secondary modality for anatomic and blood flow imaging of CHD patients whose diagnosis remain elusive after echocardiography (1-4).
The technical challenges related to the use of CMR in CHD patients can be summarized as: the anatomical structures to be visualized are highly complex and individualized (1). This complexity is heightened for the pediatric age group due to their relatively smaller organs, which requires MR acquisitions with high spatial resolution that signifies the choice for the coil and field strength in pediatric MRI (5,6). For adults with CHD, previous surgical interventions may result in unique anatomy for each individual patient (2). Faster heart rates in pediatric patients necessitate high-speed image acquisition particularly in contrast-enhanced MRI (3). Patient cooperation during the extended scan time is needed to avoid image blurring and artifacts caused by physiological motion. Patients with advanced cardiopulmonary disease as well as children may have problems with long breath-holds. This is even more challenging for toddlers and infants due to their higher cardiac and respiratory rates. Therefore, anesthesia is necessary in many situations (7-10).
Since complex CHD is usually associated with highly variable anatomical cardiac and vascular anatomic abnormalities, the validity of routine techniques for evaluating the cardiac function and deriving the heart chamber volumes and ejection fractions may need to be revisited for this group of patients. Precise chamber segmentation and accurate 3D reconstruction are two main steps toward calculation of chamber volumes. Consequently, devising more intelligent and CHD-specific segmentation and reconstruction methods is a clinical unmet need (11).
During the past few years, advancements in artificial intelligence (AI) have begun to translate into CMR aiming to overcome the existing challenges for scanning CHD patients and analyzing their data. AI has enabled faster and more robust CMR scans with improved image quality. These improvements can be categorized as: (I) image acquisition methods; (II) reconstruction techniques; and (III) post-processing algorithms. This review article is mainly focused on these three aspects and finally wraps up with the current clinical unmet needs and where the field is heading to.
AI in a nutshell
Recent advancements in computer science, more powerful computational platforms and the extensive availability of data have provided unprecedented opportunities for researchers to develop AI tools to better understand and harness complex processes. Indeed, AI has been considered one of the most promising tools in multiple aspects of medical imaging, from image acquisition and processing to aided reporting, follow-up planning, data storage, data mining, among others.
Taken as a whole, AI is a broad term referring to a field of computer science dedicated to the study of intelligent agents that mimics the cognitive functions, such as learning (gathering information and rules for a specific task), reasoning (using rules to reach conclusion) and self-correction. More recently, AI is branching off into a wide variety of subfields and techniques.
Machine learning (ML) is the main ingredient of most of the AI systems. ML enables extraction of underlying patterns governing the systems of interest with the help of data through mathematical procedures, rather than by explicit programming (12).
At the present time, artificial neural networks (ANNs) are among the most popular regression and classification class of algorithms in the realm of ML.
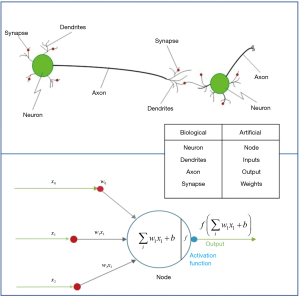
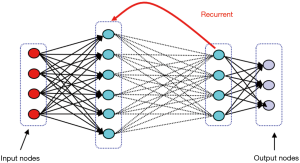
They are inspired by the human brain’s architecture of neurons and synapses (Figure 1). The popularity of ANNs is mainly due to the flexibility of modeling any complicated functional structure, especially non-linear systems. ANNs consist of sets of interconnected nodes (neurons) stacked in consecutive layers. Each node simply contains a mathematical function that transforms its input—a set of values representing features—to an output and sends it to the next layer through a weighted edge. An activation node finally transforms all the node responses to single out through a nonlinear function (13,14). The activation function is the key component of neural networks, which differentiates neural networks from a linear classifier. The performance of ANNs improves as the amount of input data and complexity of network increases. However, due to intensive computational barriers, training very large ANNs was not practical until a decade ago.
The training process is by adjusting the weights and biases of each node. Modern neural networks with millions of parameters are trained via an optimization algorithm such as gradient descent (15). In each iteration, a loss function between predictions, computed from a given input (forward propagation), and the target class, is quantitatively evaluated. Then, all randomly initialized parameters of the network are updated by small increments in the direction that minimizes the loss, a process called back-propagation.
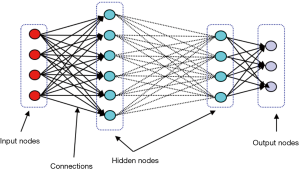
With considerable growth in computational power, deep neural network (DNN), which is basically ANN with large numbers of stacked hidden layers and neurons, has emerged (Figure 2). Since the state of layers between the first layer (input) and the last layer (output) does not correspond to observable data, they are called hidden layers (16).
Due to the ability of handling large amount of data with complex network structures, DNNs are of paramount significance in the analysis of non-numerical data structures such as image processing, speech recognition, and natural language processing (17). Among various DNNs, convolutional neural networks (CNNs), and recurrent neural networks (RNNs) are the most popular in image and video processing tasks.
CNNs
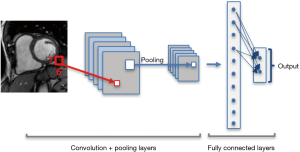
CNNs consist of special type of layers, so called convolutional layers. Each convolution operation is in fact a filter function that combines the information of neighboring inputs using learnable parameters (kernels). This type of operation is particularly advantageous in finding local patterns such as edges, shapes, lines or other visual elements in images. The convolutional layer applies multiple filters and generates multiple feature maps. In addition to convolution, other types of layers such as pooling and fully connected layers are used on CNNs (13). Pooling layers are used to capture an increasingly larger field of view, by reducing feature maps. Propagation of only the maximum or average activation, through a layer of max or average pooling, leads to lower sensitivity to small shifts or distortions of the target object in extracted feature maps (Figure 3).
Krizhevsky et al. (18) were among the first to explore much deeper convolutional networks, by proposing an 8-layer model, so called AlexNet, which competed and won the ImageNet competition in 2012. The other famous architecture, ResNet (19), whose ResNet-blocks only learn the residuals that are close to the identity function, was introduced in 2015. Using this trick, deeper models can be efficiently trained.
More recently, scientists have introduced fully convolutional neural network (FCN) (20), which is a normal CNN, except that the fully connected layers are converted to one or more convolution layers with a large “receptive field”. FCNs aim to capture the rough estimation of the locations of elements and overall context in an image. FCNs can efficiently be trained end-to-end and pixels-to-pixels for tasks like semantic segmentation.
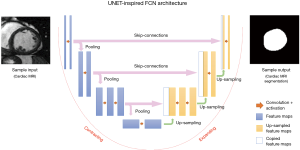
Many widely-used FCNs are inspired by the well-known U-net architecture (21), comprising a ‘regular’ FCN followed by an up-sampling part where ‘up’-convolutions are used to increase the image size, combined with so called skip-connections to directly connect opposing contracting and expanding convolutional layers. A schematic of these architectures is shown in Figure 4.
Recurrent neural networks
Standard neural networks rely on the assumption of independent training and test examples. In processing of video frames, audio, and words pulled from sentences, independent assumption fails (22).
RNNs allow network architectures with cycling and backward arcs to previous layer. This architecture is particularly useful for data with a time dependency between inputs, such as time series, and videos commonly used in medical imaging. Similar to CNNs, RNNs have deferent architecture depending on the assigned task, such as recursive, fully connected, and bidirectional layers (Figure 5) (23).
Generative adversarial neural networks
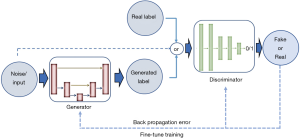
Aside from discriminative neural networks, more recently, attention has been given to generative adversarial networks (GANs). GANs are made of a pair of networks, trained simultaneously in competition with each other. One neural network, called the generator, generates new data instances, while the other, the discriminator, evaluates them and decides whether the generated data belongs to the actual training dataset or not (Figure 6).
Goodfellow et al. (24) introduced the exceptional ability of the GANs to mimic data distributions, which opens the possibility to bridge the gap between learning and synthesis. In follow-up work, Alex Radford (25) proposed an extension of GANs to address the instability issue of GANs, by converting the fully connected networks to CNNs as deep convolutional GANs (DC-GANs). Both generator and discriminator are CNNs, and all the pooling layers are replaced by strided convolutions. DC-GANs use batch normalization and ReLU (26) activation, which makes the training stable in most settings.
Due to the GANs’ excellent performance in generation of realistic-looking images, a number of articles track the recent advancements of GANs (27-29). The main reasons behind this success are the inherent advantage of being an unsupervised training method to obtain pieces of information over data (30), as well as the significant performance in the extraction of visual features by discovering the high dimensional underlying distribution of the data.
Deep learning for image acquisition
CMR involves the acquisition of cross-sectional images in standard views aligned with the heart axes. Thus, expert medical imaging technologists with detailed knowledge of cardiac anatomy are required to prescribe appropriate imaging planes with time-efficient and reproducible planning. To ensure that the heart is at the isocenter of the magnet, the technologist needs to identify the heart location from a few localizer images as one of the first steps in CMR. Therefore, automated positioning and view planning are considered important steps towards planning for automatic acquisition and post processing analysis (31). Nevertheless, due to diverse characteristics of patients’ anatomical features, automatic localization of the heart still remains an open challenge.
Conventional methods for automatic localization are mainly location-based, time-based or shape-based. In the first group, the heart is assumed to be in the center of the image (32), which does not consider the variability of patients’ anatomy and MR slices. The assumption in time-based techniques is that the only moving object in the image is the heart (33). This assumption may lead to low sensitivity, considering the other moving organs such as lungs, and the possibility of motion artifacts. Finally, the shape-based methods assume a circular shape for the left ventricle (LV) (34,35). All these assumptions usually introduce error, particularly in case of patients with extreme variabilities in their anatomy, such as CHD patients (31).
Algorithms based on ML have been employed for organ detection and localization of different parts of the body in various imaging modalities (36-38). As one of the primary machine-learning based localization methods for CMR, Zheng et al. (39) used marginal space learning to detect the LV in CMR 2D long-axis view. They also detected several important LV landmarks, such as two annulus points on the mitral valve and the apex. However, their method could not be extended to 3D or 4D images due to exponential increase in dimension of the parameter space. Lu et al. (40) used probabilistic boosting trees and marginal space learning to estimate the LV position and boundaries in the mid-ventricular short axis slice. They have used their findings for automated view planning for CMR acquisition.
More recently, the challenge of cardiac localization has been vastly tackled, due to advances in ML algorithms and in particular powerful deep learning. Kabani et al. (41) proposed a CNN architecture that treats the problem as a classification task in which pixels are classified as background or inside the bounding box. Avendi et al. (42) developed the automatic LV detection using convolutional networks to reduce the complexity of their segmentation task. They took advantage of auto-encoders to pre-train their CNN to overcome the limited number of training dataset. Other researchers utilized CNN for this purpose, which is based on exhaustive scanning of the input image searching for the LV or the heart anatomical points, without having prior knowledge about the position, the shape or the object’s motion profile (31).
The latest techniques for localization and view planning of CMR are applications of deep reinforcement learning (RL). RL is performed by interacting with an environment instead of using a set of labeled examples. Alansary et al. (43,44) proposed an RL-based approach for fully automatic view plane detection from 3D CMR data. Their model involves a complex search strategy with hierarchical action steps.
Presently, automated localization and pose detection of the heart from CMR can be streamlined using deep-learning. Since the long scan time is one of the shortcomings of MR imaging, particularly for pediatric and ACHD patients, automated view planning is the first step toward reduction of the scan time for CMR acquisition.
Deep learning for image reconstruction
Reducing the MRI scan time not only leads to higher patient satisfaction, but also helps minimizing the motion artifacts from patient movement. Since the MRI scan time is almost proportional to the number of time-consuming phase-encoding steps in k-space, under-sampling seems necessary (45). Compressed sensing MRI and Parallel MRI are some of the techniques that have been used to expedite MRI imaging while dealing with aliasing artifacts. In compressed sensing MRI (46,47), prior information on MR images of the unmeasured k-space data is used to reduce aliasing artifacts. Parallel MRI (48,49) uses space-dependent properties of the receiver coils to reduce aliasing artifacts by installing multiple receiver coils (50).
To achieve a fully sampled MR image corresponding to the under-sampled data, an optimal reconstruction function f:x→y is needed to map highly undersampled k-space data (x) to a desired image (y). Incorporating the complex MR image manifold into the regularization term using conventional regularized least-square frameworks can be challenging. Early attempts to use ML for the reconstruction task have been traditionally based on dictionary learning (51,52). They learn dictionary elements directly from under-sampled data online, such that no reference data is required. A new optimization problem must be solved for every new reconstruction, which is computationally expensive and does not involve non-linearities.
To enhance or suppress certain filter responses and by learning non-linearities, deep learning has been recently used to reconstruct anatomic MRIs and estimate reconstruction function f. In these methods, the aim is to learn the function f:x→y using training dataset {(x(i),y(i)):i=1,···,N}. Thus, f is achieved via
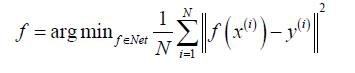
Most deep learning frameworks take advantage of highly nonlinear compressed sensing to obtain an optimal reconstruction function through the manifold constraint learned from the training set and leveraging complex prior knowledge on y. For example, Hyun et al. (45) utilized U-net (21) to provide a low-dimensional latent representation and preserve high-resolution features via concatenation in the reconstruction process. Hammernik et al. (53) proposed an MRI reconstruction approach for clinical multi-coil data based on variational methods and deep learning. They formulated MRI reconstruction as a variational model and combined this model with a gradient descent scheme, which forms a variational network structure.
A few other studies have recently discussed the application of deep learning for MRI reconstruction by leveraging patient data (54-58). These advancements in MR reconstruction and localization area, have led to significant acceleration in CMR acquisition time, as recently translated to practice by companies such as HeartVista, which is developing automated systems to reduce the scan time.
Application of deep learning in CMR post-processing
Segmentation of cardiac chambers is an important CMR post-processing technique that utilizes different image processing methods. Technically, segmentation divides an image into different parts and depicts different regions of interest. Segmentation of cardiac images provides structural information that helps characterization of different heart chambers to facilitate diagnosis of anatomic and functional disorders. Furthermore, cardiac chamber segmentation is used to calculate different clinical indices such as ejection fraction, ventricular volumes and masses. Manual segmentation of CMR images, which is currently the standard clinical practice, is time consuming and prone to inter- and intra-observer variability (34,59-61). The segmentation task is more challenging in both adult and pediatric patients with CHD due to their unique anatomical features, smaller size of heart and higher motion artifacts, as particularly seen in pediatric patients.
Classical approaches for automatic segmentation of the heart chambers can be generally classified as: region and edge-based methods (62-67), deformable methods (68-73), active appearance models (AAM) and active shape models (ASM) (74-78), and atlas models (34,76,79-81). However, these methods have many shortcomings such as low robustness and accuracy, need for extensive user interaction and sensitivity to initialization (34). A large body of literature has discussed segmentation methods for CMR images, as summarized by multiple review papers (34,82,83). The current state-of-the-art methods use supervised deep-learning-based models to outline chambers and vessels in CMR data (42,84-86). Nevertheless, due to the higher anatomical variation and fewer available training dataset for CHD patients, performance of CNNs—as the most popular models for the segmentation tasks—is still suboptimal since their performance highly depends on provided training dataset.
To address this unmet clinical need, the medical image computing and computer-assisted intervention (MICCAI) society introduced a public competition for segmentation of CHD 3D MR data in 2016 (HVSMR) (87). Yu et al. (88) won this competition by proposing a deeply-supervised 3D fractal network (3D FractalNet) for whole heart and great vessel segmentation. They employed a 3D fully convolutional architecture, organized in a self-similar recursive fractal scheme. This architecture uses multi-scale features to enhance the discrimination power by interacting sub-paths of different convolution lengths. Deep supervision (89) was used to tackle the limited number of training data, which has been shown efficient for segmentation of CHD hearts. Wolterink et al. (90) proposed another method based on 3D dilated convolutional networks. Dilated convolutional layers (91) aggregate features at multiple scales with very few parameters to avoid overfitting. Since 2016, several other methods have been proposed for automatic segmentation of CHD CMR data using CNNs (92-95). More recently, Pace et al. (96) and Rezaei et al. (97,98) employed RNNs and GANs models for whole heart segmentation of CHD patients.
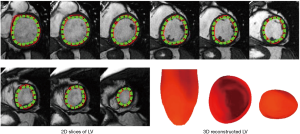
Recent attempts to automate cardiac segmentation in non-CHD patients were mainly focused on LV due to the emerging needs for assessing the left heart function. However, latest attentions to the significance of other chambers, e.g., right ventricle (RV) and left atrium (LA) in progression of the structural heart disease, necessitates approaches for quantification of multiple heart chambers. To this aim, complex anatomy of RV is very challenging to capture. Figures 7,8 illustrate the results for automated deep learning-based algorithms for LV and RV segmentation in normal CMR images.
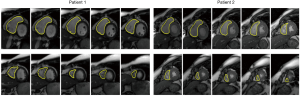
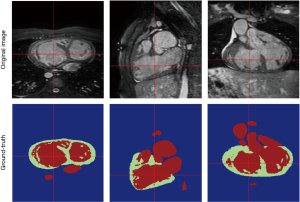
The limitation of the HVSMR dataset, the only public dataset for CHD patients, is not only the few numbers of subjects, but also there is no defined differentiation between ventricles, and each frame is divided to blood pool, myocardium and background. As a result, ventricular volumes and clinical indices cannot be calculated. Considering that one of the main reasons for segmentation is finding the volumes and indices, there is an obvious clinical unmet need for a more specified and accurate publicly-available dataset of CHD patients.
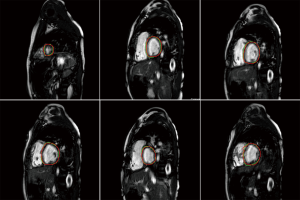
The above-mentioned studies have proved the potential of deep learning methods in CMR segmentation and the promising role of their applications in CHD patients. Yet, there are currently not many studies focused on 2D CMR data for CHD patients, which is most commonly practiced for these patients around the globe. There are only few groups working on segmentation of pediatric complex CHD MR images up to our knowledge. Figure 9 is an example of 2D MR segmentation for a complex CHD patient. Also, Figure 10 illustrates a sample image of public HVSMR dataset and the ground-truth.
Another aspect of CMR post-processing is automatic analysis of myocardial tissue characterization. This subject has been investigated in couple of very recent studies (99,100). Fahmy et al. (100) initially introduced the proof-of-concept for using deep convolutional neural networks (DCN) to automatically quantify LV mass and scar volume on CMR data with late gadolinium enhancement in patients with hypertrophic cardiomyopathy. More recently, they (99) have developed and evaluated a fully automated analysis platform for myocardial T1 mapping using FCNs. Their method automates the analysis of short-axis T1 weighted images to estimate the myocardium T1 values and was evaluated against manual T1 calculation.
AI as a diagnostic tool
Cardiac segmentation for chamber characterization is only the first step in using AI as an automated diagnostic tool for heart disease. Detection of abnormalities and disease classification are the higher level aims for use of AI in cardiac imaging. Several studies have initially utilized ML-based algorithms for automatic segmentation, computation of volumetric indices (e.g., ejection fraction) and disease classification (101-103).
In 2017, the MICCAI society released a public dataset as “Automatic Cardiac Diagnosis Challenge” (ACDC) dataset for both segmentation and diagnosis challenge. This dataset contains CMR images from 150 subjects with reference measurements and classification from two medical experts. Subjects are evenly divided into 5 classes of normal, systolic heart failure, dilated cardiomyopathy, hypertrophic cardiomyopathy, and abnormal RV, with well-defined characteristics according to physiological parameters (104).
Using this dataset, Isensee et al. (101) extracted instants and dynamic features from the segmentation maps and used an architecture with 50 multilayer perceptron (MLP) and a random forest for patients’ classification. Khened et al. (103) derived 9 features from their segmentation maps, in addition to the patients’ weight and height. They trained a 100-trees random forest classifier using those 11 features. Wolterink et al. (102) extracted 12 features from the segmentation maps, in addition to patients’ weight and height. They used a five-class random forest classifier with 1000 decision trees. Cetin et al. (105) manually extracted the contours of the cardiac structures using a semi-automatic method. Then, they derived 567 features including physiological, shape-based, intensity statistics, and various texture features. They chose the most discriminative features to prevent overfitting and used support vector machine (SVM) for classification. More recently, Snaauw et al. (106) proposed an end-to-end multi-task learning algorithm for segmentation and classification of ACDC dataset. Although their classification accuracy was lower than hand-crafted methods, they reported competitive results.
Aside from studies mentioned above, only few studies have been conducted on cardiac, disease diagnosis or classification (107,108), and to the best of our knowledge, no study has yet reported on diagnosis of CHD either in adult or pediatric patient groups.
AI challenges in CMR
Despite many promising results on AI applications in CMR, multiple challenges are on the way of translation of AI algorithms and methods into clinical practice. We have listed some of those here.
Flow analysis
One of the needed applications of AI in CMR is analysis of cardiac fluid dynamics and flow measurements. Considering the complex features of 4D flow CMR images, a potential space for AI improvement is to help analysis of complex fluid dynamics in CHD patients using 4D flow CMR.
Dataset
The performance of most AI algorithms highly depends on the quality and quantity of training data. On top of high costs for preparing large enough annotated CMR datasets, there is a considerable difference in the quality of scanned data (i.e., ones typically used for research and the ones used for clinical purposes). This heterogeneity in the quality of the data can be an obstacle for network’s generalization, and a major challenge for commercialization of AI-based systems (109). There are other data variabilities such as abnormal anatomical features, MRI machine vendors and protocols in different hospital. Therefore, developing evaluation methods to test general performance of each technique is a clinical unmet need. Accordingly, multi-center and multi-vendor studies are highly recommended to mitigate the heterogeneity of the scanned data (16,83).
The other data-related issue in medical imaging is class imbalance, which is particularly important in CHD. In most of the currently-available datasets, the majority of images are from normal subjects with very few abnormal cases. This can lead to lower accuracy of AI methods for real-life applications. Thus, development of AI platforms that can handle this class imbalance is of significant importance (110).
Data privacy
The patients’ privacy and data security are the top priority requirement when dealing with medical data. It is much more complicated and difficult to share the medical data as compared to natural images. In the US, Health Insurance Portability and Accountability Act (HIPAA) provides legal rights to patients regarding their personally-identifiable information and establish obligations for healthcare providers to protect and restrict their use with disclosure. Thus, all communications need to be secured, and all data should first be encrypted per HIPAA rules to remove any identifier. These privacy challenges are factors that can lead to trust and legal issues towards translation of AI models into clinical practice and may even negatively impact it.
IP and legal issues
Like any novel technology, there could be legal ramifications regarding the use of clinical imaging data for the commercial development of AI-based systems, since the owner of the data is not well specified. As well, there could be ambiguity in terms of intellectual property among data owners, including patients, data collectors, and algorithm developers. New regulations regarding data ownership and algorithms need to be developed to help development of these AI-based technologies.
Liability
Implementation of AI in diagnostic procedures raises the legal liability and ethical issues independent from supervision of an imaging physician. Errors that affect the diagnosis may have serious ramifications for the patients. Who or what should take the responsibility in case of an AI-generated mistake that harm a patient?
These questions have always been posed and resolved in the history of technology development. Considering vast applications of AI in human life, these concerns need to be further studied and resolved in the coming years (111).
Conclusions
CMR allows for accurate analysis of cardiac functions. In this paper, we have reviewed the state-of-the-art AI-based methods which successfully improve every steps of CMR analysis. These methods proved their power in analysis of normal or non-CHD subjects by being applied on multiple publicly available datasets. However, only one public dataset is currently available for CHD patients, which has substantial limitations. Considering the significance of early diagnosis and disease management for CHD patients and at the same time the complexity and challenges of CMR for this group of patients, the role of AI is needed to be more significantly investigated.
Acknowledgments
The authors would also like to acknowledge Saeed Karimi Bidhendi for providing the images in 9th illustration. from his work-in-progress study on automatic segmentation of tetralogy of Fallot. This work was supported by an experienced researcher award from the Alexander von Humboldt Foundation to Prof. Kheradvar.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Lawley CM, Broadhouse KM, Callaghan FM, et al. 4D flow magnetic resonance imaging: role in pediatric congenital heart disease. Asian Cardiovasc Thorac Ann 2018;26:28-37. [Crossref] [PubMed]
- Prakash A, Powell AJ, Geva T. Multimodality Noninvasive Imaging for Assessment of Congenital Heart Disease. Circ Cardiovasc Imaging 2010;3:112-25. [Crossref] [PubMed]
- Gershlick A, de Belder M, Chambers J, et al. Role of non-invasive imaging in the management of coronary artery disease: an assessment of likely change over the next 10 years. A report from the British Cardiovascular Society Working Group. Heart 2007;93:423-31. [Crossref] [PubMed]
- Partington SL, Valente AM. Cardiac magnetic resonance in adults with congenital heart disease. Methodist Debakey Cardiovasc J 2013;9:156-62. [Crossref] [PubMed]
- Ravi A. Efficacy of a Multi-Channel Array Coil for Pediatric Cardiac Magnetic Resonance Imaging. The Graduate Faculty of The University of Akron. 2008.
- Vasanawala SS, Lustig M. Advances in Pediatric Body MRI. Pediatr Radiol 2011;41:549-54. [Crossref] [PubMed]
- Driessen MM, Breur JM, Budde RP, et al. Advances in cardiac magnetic resonance imaging of congenital heart disease. Pediatr Radiol 2015;45:5-19. [Crossref] [PubMed]
- Ciet P, Tiddens HA, Wielopolski PA, et al. Magnetic resonance imaging in children: common problems and possible solutions for lung and airways imaging. Pediatr Radiol 2015;45:1901-15. [Crossref] [PubMed]
- Greil G, Tandon AA, Silva Vieira M, et al. 3D Whole Heart Imaging for Congenital Heart Disease. Front Pediatr 2017;5:36. [Crossref] [PubMed]
- Chan FP, Hanneman K. Computed tomography and magnetic resonance imaging in neonates with congenital cardiovascular disease. Semin Ultrasound CT MR 2015;36:146-60. [Crossref] [PubMed]
- Niwa K, Uchishiba M, Aotsuka H, et al. Measurement of ventricular volumes by cine magnetic resonance imaging in complex congenital heart disease with morphologically abnormal ventricles. Am Heart J 1996;131:567-75. [Crossref] [PubMed]
- Erickson BJ, Korfiatis P, Akkus Z, et al. Machine Learning for Medical Imaging. RadioGraphics 2017;37:505-15. [Crossref] [PubMed]
- Al'Aref SJ, Anchouche K, Singh G, et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J 2019;40:1975-86. [Crossref] [PubMed]
- Haykin S. Neural Networks: A Comprehensive Foundation. New York: Prentice hall, 1994.
- Barzilai J, Borwein JM. Two-Point Step Size Gradient Methods. IMA J NUMER ANAL 1988;8:141-8. [Crossref]
- Lee JG, Jun S, Cho YW, et al. Deep learning in medical imaging: general overview. Korean J Radiol 2017;18:570-84. [Crossref] [PubMed]
- Srivastava S, Soman S, Rai A, et al. Deep learning for health informatics: Recent trends and future directions. In: 2017 International Conference on Advances in Computing, Communications and Informatics (ICACCI). IEEE, 2017:1665-70.
- Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems 25 (NIPS 2012) 2012:1097-105.
- He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. IEEE, 2016:770-8.
- Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. IEEE, 2015:3431-40.
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, et al. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Springer, 2015:234-41.
- Lipton ZC, Berkowitz J, Elkan C. A critical review of recurrent neural networks for sequence learning. 2015. arXiv:1506.00019.
- Schuster M, Paliwal KK. Bidirectional recurrent neural networks. IEEE Trans Signal Process 1997;45:2673-81. [Crossref]
- Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. In: Advances in Neural Information Processing Systems 27 (NIPS 2014). 2014:2672-80.
- Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversari.al networks. 2015. arXiv:1511.06434.
- Rectifier (neural networks). Wikipedia. 2019. Available online: https://en.wikipedia.org/wiki/Rectifier_(neural_networks)
- Kazeminia S, Baur C, Kuijper A, et al. GANs for Med Image Anal. 2018. arXiv:1809.06222.
- Creswell A, White T, Dumoulin V, et al. Generative adversarial networks: An overview. IEEE Signal Process Mag 2018;35:53-65. [Crossref]
- Hong Y, Hwang U, Yoo J, et al. How Generative Adversarial Nets and its variants Work: An Overview. 2017. arXiv:1711.05914.
- Isola P, Zhu JY, Zhou T, et al. Image-to-image translation with conditional adversarial networks. arXiv:1611.07004.
- Emad O, Yassine IA, Fahmy AS. Automatic localization of the left ventricle in cardiac MRI images using deep learning. In: 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE, 2015:683-6.
- O'Brien SP, Ghita O, Whelan PF. A Novel Model-Based 3D+Time Left Ventricular Segmentation Technique. IEEE Trans Med Imaging 2011;30:461-74. [Crossref] [PubMed]
- Hoffmann R, Bertelshofer F, Siegl C, et al. Automated Heart Localization in Cardiac Cine MR Data. In: Tolxdorff T, Deserno TM, Handels H, et al. editors. Bildverarbeitung für die Medizin 2016. Berlin: Springer Vieweg, 2016:116-21.
- Petitjean C, Dacher JN. A review of segmentation methods in short axis cardiac MR images. Med Image Anal 2011;15:169-84. [Crossref] [PubMed]
- Zhong L, Zhang JM, Zhao X, et al. Automatic Localization of the Left Ventricle from Cardiac Cine Magnetic Resonance Imaging: A New Spectrum-Based Computer-Aided Tool. PLoS One 2014;9:e92382. [Crossref] [PubMed]
- Seifert S, Barbu A, Zhou SK, et al. Hierarchical Parsing and Semantic Navigation of Full Body CT Data. In: Pluim JPW, Dawant BM. editors. Proceedings of SPIE 7259, Medical Imaging 2009: Image Processing. 2009:725902.
- Zheng Y, Georgescu B, Ling H, et al. Constrained marginal space learning for efficient 3D anatomical structure detection in medical images. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE, 2009:194-201.
- Criminisi A, Shotton J, Robertson D, et al. Regression forests for efficient anatomy detection and localization in CT studies. In: Menze B, Langs G, Tu Z, et al. editors. International MICCAI Workshop on Medical Computer Vision. Medical Computer Vision. Recognition Techniques and Applications in Medical Imaging. Berlin: Springer, 2010:106-17.
- Zheng Y, Lu X, Georgescu B, et al., editors. Robust object detection using marginal space learning and ranking-based multi-detector aggregation: Application to left ventricle detection in 2D MRI images. 2009 IEEE Conference on Computer Vision and Pattern Recognition. 2009. Available online: https://ieeexplore.ieee.org/document/5206808
- Lu X, Jolly MP, Georgescu B, et al. Automatic View Planning for Cardiac MRI Acquisition. In: Fichtinger G, Martel A, Peters T. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2011. Springer, 2011:479-86.
- Kabani A, El-Sakka MR. Object Detection and Localization Using Deep Convolutional Networks with Softmax Activation and Multi-class Log Loss. In: Campilho A, Karray F. editors. Image Analysis and Recognition. Springer International Publishing, 2016:358-66.
- Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal 2016;30:108-19. [Crossref] [PubMed]
- Alansary A, Folgoc LL, Vaillant G, et al. Automatic View Planning with Multi-scale Deep Reinforcement Learning Agents. 2018. arXiv:180603228.
- Alansary A, Oktay O, Li Y, et al. Evaluating reinforcement learning agents for anatomical landmark detection. Med Image Anal 2019;53:156-64. [Crossref] [PubMed]
- Hyun CM, Kim HP, Lee SM, et al. Deep learning for undersampled MRI reconstruction. Phys Med Biol 2018;63:135007. [Crossref] [PubMed]
- Lustig M, Donoho DL, Santos JM, et al. Compressed sensing MRI. IEEE Signal Process Mag 2008;25:72-82. [Crossref]
- Donoho DL. Compressed sensing. IEEE Trans Inf Theory 2006;52:1289-306. [Crossref]
- Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002;47:1202-10. [Crossref] [PubMed]
- Pruessmann KP, Weiger M, Scheidegger MB, et al. SENSE: sensitivity encoding for fast MRI. Magn Reson Med 1999;42:952-62. [Crossref] [PubMed]
- Lee D, Yoo J, Ye JC. Deep artifact learning for compressed sensing and parallel MRI. 2017. arXiv:1703.01120.
- Caballero J, Price AN, Rueckert D, et al. Dictionary learning and time sparsity for dynamic MR data reconstruction. IEEE Trans Med Imaging 2014;33:979-94. [Crossref] [PubMed]
- Ravishankar S, Bresler Y. MR image reconstruction from highly undersampled k-space data by dictionary learning. IEEE Trans Med Imaging 2011;30:1028. [Crossref] [PubMed]
- Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med 2018;79:3055-71. [Crossref] [PubMed]
- Lee D, Yoo J, Ye JC, Deep residual learning for compressed sensing MRI. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017). IEEE, 2017:15-8.
- Majumdar A. Real-time dynamic mri reconstruction using stacked denoising autoencoder. 2015. arXiv:1503.06383.
- Zhu B, Liu JZ, Cauley SF, et al. Image reconstruction by domain-transform manifold learning. Nature 2018;555:487. [Crossref] [PubMed]
- Wang G, Ye JC, Mueller K, et al. Image reconstruction is a new frontier of machine learning. IEEE Trans Med Imaging 2018;37:1289-96. [Crossref] [PubMed]
- Mardani M, Gong E, Cheng JY, et al. Deep Generative Adversarial Neural Networks for Compressive Sensing MRI. IEEE Trans Med Imaging 2019;38:167-79. [Crossref] [PubMed]
- Tavakoli V, Amini AA. A survey of shaped-based registration and segmentation techniques for cardiac images. Comput Vis Image Underst 2013;117:966-89. [Crossref]
- Heimann T, Meinzer HP. Statistical shape models for 3D medical image segmentation: a review. Med Image Anal 2009;13:543-63. [Crossref] [PubMed]
- Suinesiaputra A, Cowan BR, Al-Agamy AO, et al. A collaborative resource to build consensus for automated left ventricular segmentation of cardiac MR images. Med Image Anal 2014;18:50-62. [Crossref] [PubMed]
- Jolly MP, Xue H, Grady L, et al. Combining registration and minimum surfaces for the segmentation of the left ventricle in cardiac cine MR images. Med Image Comput Comput Assist Interv 2009;12:910-8. [PubMed]
- Jolly M. Fully automatic left ventricle segmentation in cardiac cine MR images using registration and minimum surfaces. The MIDAS Journal-Cardiac MR Left Ventricle Segmentation Challenge. Available online: http://hdl.handle.net/10380/3114
- Lee H-Y, Codella N, Cham M, et al. Left ventricle segmentation using graph searching on intensity and gradient and a priori knowledge (lvGIGA) for short-axis cardiac magnetic resonance imaging. J Magn Reson Imaging 2008;28:1393-401. [Crossref] [PubMed]
- Lin X, Cowan BR, Young AA. Automated detection of left ventricle in 4D MR images: experience from a large study. Med Image Comput Comput Assist Interv 2006;9:728-35. [PubMed]
- Uzümcü M, van der Geest RJ, Swingen C, et al. Time continuous tracking and segmentation of cardiovascular magnetic resonance images using multidimensional dynamic programming. Invest Radiol 2006;41:52-62. [Crossref] [PubMed]
- Codella NC, Weinsaft JW, Cham MD, et al. Left Ventricle: Automated Segmentation by Using Myocardial Effusion Threshold Reduction and Intravoxel Computation at MR Imaging 1. Radiology 2008;248:1004-12. [Crossref] [PubMed]
- Queirós S, Barbosa D, Heyde B, et al. Fast automatic myocardial segmentation in 4D cine CMR datasets. Med Image Anal 2014;18:1115-31. [Crossref] [PubMed]
- Paragios N. A level set approach for shape-driven segmentation and tracking of the left ventricle. IEEE Trans Med Imaging 2003;22:773-6. [Crossref] [PubMed]
- Pluempitiwiriyawej C, Moura JMF, Wu YJL, et al. STACS: new active contour scheme for cardiac MR image segmentation. IEEE Trans Med Imaging 2005;24:593-603. [Crossref] [PubMed]
- Li C, Xu C, Gui C, et al. Distance regularized level set evolution and its application to image segmentation. IEEE Trans Image Process 2010;19:3243-54. [Crossref] [PubMed]
- Barbosa D, Dietenbeck T, Schaerer J, et al. B-Spline Explicit Active Surfaces: An Efficient Framework for Real-Time 3-D Region-Based Segmentation. IEEE Trans Image Process 2012;21:241-51. [Crossref] [PubMed]
- Lee HY, Codella NC, Cham MD, et al. Automatic Left Ventricle Segmentation Using Iterative Thresholding and an Active Contour Model With Adaptation on Short-Axis Cardiac MRI. IEEE Trans Biomed Eng 2010;57:905-13. [Crossref] [PubMed]
- Suinesiaputra A, Frangi AF, Kaandorp T, et al. Automated Detection of Regional Wall Motion Abnormalities Based on a Statistical Model Applied to Multislice Short-Axis Cardiac MR Images. IEEE Trans Med Imaging 2009;28:595-607. [Crossref] [PubMed]
- Koikkalainen J, Tolli T, Lauerma K, et al. Methods of Artificial Enlargement of the Training Set for Statistical Shape Models. IEEE Trans Med Imaging 2008;27:1643-54. [Crossref] [PubMed]
- Zhang H, Wahle A, Johnson RK, et al. 4-D cardiac MR image analysis: left and right ventricular morphology and function. IEEE Trans Med Imaging 2010;29:350-64. [Crossref] [PubMed]
- Van Assen HC, Danilouchkine MG, Frangi AF, et al. SPASM: a 3D-ASM for segmentation of sparse and arbitrarily oriented cardiac MRI data. Med Image Anal 2006;10:286-303. [Crossref] [PubMed]
- Mansi T, Voigt I, Leonardi B, et al. A statistical model for quantification and prediction of cardiac remodelling: Application to tetralogy of fallot. IEEE Trans Med Imaging 2011;30:1605-16. [Crossref] [PubMed]
- Zhuang X, Rhode K, Arridge S, et al. An atlas-based segmentation propagation framework locally affine registration--application to automatic whole heart segmentation. Med Image Comput Comput Assist Interv 2008;11:425-33. [PubMed]
- Isgum I, Staring M, Rutten A, et al. Multi-Atlas-Based Segmentation With Local Decision Fusion -- Application to Cardiac and Aortic Segmentation in CT Scans. IEEE Trans Med Imaging 2009;28:1000-10. [Crossref] [PubMed]
- Lorenzo-Valdés M, Sanchez-Ortiz GI, Elkington AG, et al. Segmentation of 4D cardiac MR images using a probabilistic atlas and the EM algorithm. Med Image Anal 2004;8:255-65. [Crossref] [PubMed]
- Peng P, Lekadir K, Gooya A, et al. A review of heart chamber segmentation for structural and functional analysis using cardiac magnetic resonance imaging. MAGMA 2016;29:155-95. [Crossref] [PubMed]
- Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys 2019;29:102-27. [Crossref] [PubMed]
- Avendi MR, Kheradvar A, Jafarkhani H. Automatic segmentation of the right ventricle from cardiac MRI using a learning-based approach. Magn Reson Med 2017;78:2439-48. [Crossref] [PubMed]
- Wang C, Smedby Ö. Automatic Whole Heart Segmentation Using Deep Learning and Shape Context. In: Mansi T, McLeod K, Pop M, et al. editors. International Workshop on Statistical Atlases and Computational Models of the Heart. STACOM 2017: Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges. Springer, 2017:242-9.
- Payer C, Štern D, Bischof H, et al. Multi-label whole heart segmentation using CNNs and anatomical label configurations. In: Young A, Bernard O. editors. STACOM 2017: Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges. Springer, 2017:190-8.
- Pace DF, Dalca AV, Geva T, et al. Interactive Whole-Heart Segmentation in Congenital Heart Disease. Med Image Comput Comput Assist Interv 2015;9351:80-8. [PubMed]
- Yu L, Yang X, Qin J, et al. 3D FractalNet: Dense Volumetric Segmentation for Cardiovascular MRI Volumes. In: Zuluaga MA, Bhatia K, Kainz B, et al. editors. Reconstruction, Segmentation, and Analysis of Medical Images. Cham: Springer International Publishing, 2017.
- Dou Q, Chen H, Jin Y, et al. 3D deeply supervised network for automatic liver segmentation from CT volumes. 2016. arXiv:1607.00582.
- Wolterink JM, Leiner T, Viergever MA, et al. Dilated Convolutional Neural Networks for Cardiovascular MR Segmentation in Congenital Heart Disease. 2017. arXiv:1704.03669.
- Yu F, Koltun V. Multi-scale context aggregation by dilated convolutions. 2015. arXiv:1511.07122.
- Li J, Zhang R, Shi L, et al. Automatic Whole-Heart Segmentation in Congenital Heart Disease Using Deeply-Supervised 3D FCN. In: Zuluaga MA, Bhatia K, Kainz B, et al. editors. Reconstruction, Segmentation, and Analysis of Medical Images. Springer, 2017:111-8.
- Mukhopadhyay A. Total Variation Random Forest: Fully Automatic MRI Segmentation in Congenital Heart Diseases. In: Zuluaga MA, Bhatia K, Kainz B, et al. editors. Reconstruction, Segmentation, and Analysis of Medical Images. Springer, 2017:165-71.
- Tziritas G. Fully-Automatic Segmentation of Cardiac Images Using 3-D MRF Model Optimization and Substructures Tracking. In: Zuluaga MA, Bhatia K, Kainz B, et al. editors. Reconstruction, Segmentation, and Analysis of Medical Images. Springer, 2017:126-36.
- Yu L, Cheng J-Z, Dou Q, et al. editors. Automatic 3D Cardiovascular MR Segmentation with Densely-Connected Volumetric ConvNets. In: Descoteaux M, Maier-Hein L, Franz A, et al. editors. Medical Image Computing and Computer-Assisted Intervention − MICCAI 2017. Cham: Springer International Publishing, 2017:287-95.
- Pace DF, Dalca AV, Brosch T, et al. Iterative Segmentation from Limited Training Data: Applications to Congenital Heart Disease. 2018. arXiv:1708.00573.
- Rezaei M, Yang H, Meinel C. Recurrent generative adversarial network for learning imbalanced medical image semantic segmentation. Multimed Tools Appl 2019. doi: 10.1007/s11042-019-7305-1. [Crossref]
- Rezaei M, Yang H, Meinel C. Whole Heart and Great Vessel Segmentation with Context-aware of Generative Adversarial Networks. In: Maier A, Deserno TM, Handels H, et al. editors. Bildverarbeitung für die Medizin 2018. Berlin: Springer, 2018:353-8.
- Fahmy AS, El-Rewaidy H, Nezafat M, et al. Automated analysis of cardiovascular magnetic resonance myocardial native T 1 mapping images using fully convolutional neural networks. J Cardiovasc Magn Reson 2019;21:7. [Crossref] [PubMed]
- Fahmy AS, Rausch J, Neisius U, et al. Automated cardiac MR scar quantification in hypertrophic cardiomyopathy using deep convolutional neural networks. JACC Cardiovasc Imaging 2018;11:1917-8. [Crossref] [PubMed]
- Isensee F, Jaeger PF, Full PM, et al. Automatic Cardiac Disease Assessment on cine-MRI via Time-Series Segmentation and Domain Specific Features. In: In: Pop M, Sermesant M, Jodoin PM, et al. editors. Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges. Cham: Springer International Publishing, 2017:120-9.
- Wolterink JM, Leiner T, Viergever MA, et al. Automatic segmentation and disease classification using cardiac cine MR images. In: Pop M, Sermesant M, Jodoin PM, et al. editors. Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges. Springer, 2017:101-10.
- Khened M, Alex V, Krishnamurthi G. Densely connected fully convolutional network for short-axis cardiac cine mr image segmentation and heart diagnosis using random forest. In: Pop M, Sermesant M, Jodoin PM, et al. editors. Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges. Springer, 2017:140-50.
- Bernard O, Lalande A, Zotti C, et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans Med Imaging 2018;37:2514-25. [Crossref] [PubMed]
- Cetin I, Sanroma G, Petersen SE, et al. A radiomics approach to computer-aided diagnosis with cardiac cine-MRI. In: Pop M, Sermesant M, Jodoin PM, et al. editors. Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges. Springer, 2017:82-90.
- Snaauw G, Gong D, Maicas G, et al. End-to-End Diagnosis and Segmentation Learning from Cardiac Magnetic Resonance Imaging. 2018. arXiv:1810.10117.
- Chang Y, Song B, Jung C, et al., editors. Automatic Segmentation and Cardiopathy Classification in Cardiac Mri Images Based on Deep Neural Networks. 2016 Fourth International Conference on Parallel, Distributed and Grid Computing (PDGC). 2016. Available online: https://ieeexplore.ieee.org/abstract/document/8461261
- Sharma L, Gupta G, Jaiswal V, editors. Classification and development of tool for heart diseases (MRI images) using machine learning. 2016 Fourth International Conference on Parallel, Distributed and Grid Computing (PDGC). 2016. Available online: https://ieeexplore.ieee.org/document/7913149
- Zech JR, Badgeley MA, Liu M, et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med 2018;15:e1002683. [Crossref] [PubMed]
- Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- Razzak MI, Naz S, Zaib A. Deep learning for medical image processing: Overview, challenges and the future. In: Dey N, Ashour AS, Borra S. Classification in BioApps. Springer, 2018:323-50.