
N6-methyladenosine (m6A) RNA modification in the pathophysiology of heart failure: a narrative review
Introduction
Heart failure (HF) is a syndrome characterized by structural and/or functional abnormalities of the heart, resulting in elevated intracardiac pressures and/or inadequate cardiac output at rest and/or during exercise (1). Approximately 6 million adults (≥20 years) in the United States experienced HF from 2015 to 2018 (2). Nationally, HF care costs were estimated at $31 billion in 2012, including costs for healthcare services, HF medications, and work absences (3). Therefore, investigation of HF pathogenesis is vital to enhance understanding of the underlying mechanisms and improve therapeutics, clinical outcomes, and care costs.
The failing heart undergoes structural and functional alterations through substantial transcriptional reprogramming at the messenger RNA (mRNA) level (4). Anormal modifications of RNA have been identified in cardiovascular disease and have attracted attention for improving understanding of the mechanisms underlying cardiovascular disease development (5,6). Epigenetics can also impact gene expression and function without altering the base sequence of DNA. Moreover, these effects are reversible, heritable, and influenced by the external environment (7). Epigenetic regulatory mechanisms are instrumental in cardiovascular development, tissue homeostasis, and disease progression (8). More than 100 RNA modifications decorate the chemical compound and the topological properties of ribonucleotides, thereby executing their biological functions through post-transcriptional regulation. In cardiovascular diseases, various RNA modifications, including N6-adenosine methylation (m6A), 5-methyl-cytidine (m5C), 2'-O-ribose-methylation (Nm), pseudouridine (Ψ), N7-methylguanosine (m7G), and N1-adenosine methylation (m1A), have been found in transfer RNA (tRNA), ribosomal RNA (rRNA), mRNA, and other noncoding RNAs. These modifications can function as novel mechanisms in metabolic syndrome, HF, coronary heart disease, and hypertension (9). As an abundant internal mRNA modification, m6A is involved in the regulation of RNA splicing, localization, translation, and decay (10,11). Generally, m6A modifications are embedded within the conserved sequence 5'-RRACU-3' (where R = A or G; the methylated adenosine residue is underscored), and predominantly occur at the beginning of the 3' untranslated region (3'-UTR) near the translation-termination codon (12). m6A can regulate gene expression and participates in various cellular processes, such as cell renewal, apoptosis, immunity, differentiation, and cancer-cell invasion. Abnormal regulation of m6A may cause cancer and cardiovascular disease (13). Recent studies demonstrated that m6A modification is involved in risk factors for HF and the pathophysiological processes leading to HF, such as ischemic heart disease (14), cardiac hypertrophy (15), and ischemia/reperfusion-induced cardiomyopathy (16). However, there are limited studies providing definitive conclusions on the role of m6A in the progression of HF. This review elucidates the pathological processes involving m6A modifications and describes promising therapeutic targets related to m6A modifications in HF. We present the following article in accordance with the Narrative Review reporting checklist (available at https://cdt.amegroups.com/article/view/10.21037/cdt-22-277/rc).
Methods
Searching strategy
We searched PubMed, Web of science and Medline for articles in English related to this topic using the following searching strategy:
- #1 “Heart failure” OR “heart dysfunction”;
- #2 “myocardium fibrosis” OR “myocardium hypertrophy” OR “cardiac cell apoptosis” OR “myocardium ischemic reperfusion injury”;
- #3 “N6-adenosine methylation” OR “m6A”;
- #4 #1 AND #3;
- #5 #2 AND #3.
We put the searching results into the Endnote software to make further filtration and select those related articles published in recent 10 years [2012–2022] for further full text analysis. The details of searching were illustrated in Table 1.
Table 1
| Items | Specification |
|---|---|
| Date of search | 2021.10.25–2022.1.15 |
| Databases and other sources searched | PubMed, Web of Science and Medline |
| Search terms used | #1 “Heart failure” OR “heart dysfunction” |
| #2 “myocardium fibrosis” OR “myocardium hypertrophy” OR “cardiac cell apoptosis” OR “myocardium ischemic reperfusion injury” | |
| #3 “N6-adenosine methylation” OR “m6A” | |
| #4 #1 AND #3 | |
| #5 #2 AND #3 | |
| Timeframe | 2012–2022 |
| Inclusion and exclusion criteria | Inclusion criteria: research work related to the m6A and the pathophysiological process of heart failure. The language is English. The type of research work is cell or in vivo experiment |
| Selection process | We put the searching results into the Endnote software to make further filtration and select those related articles published in recent 10 years [2012–2022] for further full text analysis. Two researchers (Tongyu Wang and Sihan Liu) finished the selection process independently |
Biological function of m6A
m6A participates in the metabolism of RNA
In eukaryotes, control of mRNA translation and degradation is critical for spatio-temporal and specific regulation of gene expression. There are over 100 types of RNA modification, and although these are essential for the function of RNA, the significance of most RNA chemical modifications has not been fully elucidated. Most of RNA’s existence occurs with “temporary” modifications, although modifications on RNA are covalent bonds and immutable (17). More than 60% of chemical RNA modifications are methylations and the m6A modification is highly abundant and conserved in mammals. The m6A modification is a dynamic and reversible process under the action of its regulatory molecules. Due to the steric hindrance effect of RNA methylation, stability of the hydrogen bond is reduced and the thermal stability of the m6A modified transcript is weakened (18).
Regulatory molecules of m6A can be classified into three main categories: readers, writers, and erasers. Writers, primarily methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14), generally perform RNA methylation modifications using the SAM binding motif and the cofactor Wilms tumor 1-associating protein (WTAP) (19). Erasers include fat mass and obesity-associated gene (FTO) and AlkB homolog 5 (ALKBH5). FTO belongs to the AlkB protein family—which contains a double-stranded, beta-helix (DSBH) fold homologous to those of Fe(II) and 2-oxoglutarate (2OG) oxygenases—and performs its m6A eraser function via an oxidative demethylation reaction (20,21). Readers mainly comprise YTH domain family (YTHDF)1, YTHDF2, YTHDF3, YTH domain containing (YTHDC)1, and YTHDC2, which all have the same YT521B homology (YTH) domain and the ability to bind to m6A-modified RNA transcripts (21). Binding between m6A readers and m6A-modified RNA affects subsequent metabolic activities of RNA. Meanwhile, readers also contain insulin-like growth factor 2 mRNA-binding protein (IGF2BP)1/2/3 and three heterogenous nuclear ribonucleoproteins (hnRNPs)—hnRNPC, hnRNPGm, and hnRNPA2B1—which increase the stability of the RNA transcript and promote the maturation of microRNA, respectively (22). Therefore, m6A modification differs from the traditional cognition of post-transcriptional mRNA modifications in that m6A modification can regulate the stability, output, splicing, degradation, and translation processes of RNA, and interact with other epigenetic modifications.
Subcellular localization analysis of the m6A-coding protein YTHDF2 revealed colocalization with DCP1a, GW182, and DDX6 P-bodies, which are associated with intracellular RNA degradation. This suggested that YTHDF2 affects the subcellular localization of m6A-methylated transcripts, and transfer from the translation pool to the decay pool affects the translation and half-life of transcripts (23). In contrast, YTHDF1 facilitates the transfer of ribosomes to transcripts and accelerates the protein translation process (24). Immunoprecipitation studies established that YTHDF1 binds to the translation initiation complex, eukaryotic initiation factor 3 (eIF3), to affect protein translation initiation. This process is m6A-dependent and does not require the participation of eukaryotic translation initiation factor 4E (eIF4E) (25). Additionally, the influence of m6A on alternative mRNA splicing requires the nucleus m6A recognition factor YTHDC1, which combines with Src-associated in mitosis 68KD (SAM68), SC35, and serine/arginine-rich splicing factor (SRSF) to regulate alternative splicing (26,27). YTHDC1 can also bind to SRSF3 protein. SRSF3 promotes the transport of mRNA from inside the nucleus to outside by binding to nuclear RNA export factor 1 (NXF1) protein and m6A-methylated mRNA can bind to the coding protein YTHDC1. This implied that m6A indirectly causes the transport of mRNA from the nucleus to the cytoplasm (28). As for the result of YTHDC1 gene alternation, a study utilized multiple types of methods to test the results. The liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) result showed no significant change in m6A/A ratios after YTHDC1 gene knocking-out. However, the number of m6A-methylated transcripts in the nucleus observed by in situ fluorescence hybridization increased. The above changes did not significantly alter the expression of nuclear export machinery NXF1, nor its adaptor protein ALYREF. Over-expression of YTHDC1 decreased m6A-methylated mRNA transcripts in the nucleus but did not increase the number of m6A-methylated transcripts in the cytoplasm. The study also used immunoprecipitation to demonstrate that YTHDC1 binds to and interacts with the Zn-finger protein, ZCCHC8, a component of the trimeric nuclear exosome targeting complex (NEXT) (29). The RNA exosome complex controls the level of intracellular RNA in a dynamic equilibrium state with its 3'-5' degradation ability. Therefore, YTHDC1 is involved in intranuclear degradation of m6A-labeled mRNA (30).
In addition to the YTH family, recent study found that the m6A writer can also interact with eIF4A2 to facilitate gene expression in cardiac cells under hypoxic conditions via the scaffold function of nuclear cap-binding subunit 3 (NCBP3) (31). Besides, the METTL3 was also found to be involved in the RNA splicing process. Studies on the sex-lethal gene of drosophila, SXL, found that female drosophila expressed male-specific variable SXL splicing transcripts after deletion of the m6A methylase homolog METTL3, Ime4 (32). Splicing regulators such as Fl(2)D, Virilizer33, and Spenito (Nito) are homologous to the mammalian m6A methylation transferases, WTAP, KIAA1429, and RBM15/RBM15B, respectively (33,34), which suggests m6A may participate in alternative RNA splicing. Further clarification is required to determine whether m6A modification is distributed near introns or exons close to the alternative splicing site. However, it is very difficult to locate m6A in the intron region due to the rarity of intron components in cells (35). Currently, most studies indicate that m6A is enriched in flanking 5'- and 3'-splicing sites in the exon region. These regions are regulatory mRNA-splicing serine- and arginine-rich proteins. The m6A deletion factor, FTO, in the SRSF2 binding region leads to an increased level of m6A in the exon region and improves the mRNA binding capacity of SRSF2, resulting in increased expression of the target exon (36). In addition to being a recognition molecule of m6A, FTO is also involved in the RNA precursor splicing process. Immunofluorescence technology revealed that FTO is mainly distributed in the nucleus, while crosslinking-immunoprecipitation (CLIP) technology clarified that FTO preferentially binds to the intron region, and most FTO-bound regions are located at the junction between the intron and exon. These binding sites are related to the regulatory function of alternative m6A-modified alternative splicing. Subsequent analysis of the effect of deleting FTO on BRD8 alternative splicing showed that more exon-skipping events occurred after FTO deletion. By using a vector to engineer knockout cells to re-express FTO, exon skipping was reduced, implying that the regulation of m6A modifications is involved in the regulation of alternative splicing (37,38)
ALKBH5, another regulator of m6A methylation modifications, is also involved in the RNA transport process. Immunofluorescence experiments revealed that total mRNA levels in the cytoplasm increased after knockout of ALKBH5, while the distribution of rRNA did not change significantly, suggesting that ALKBH5 alteration affects the subcellular localization of RNA and has certain selectivity. ALKBH5 was previously reported to colocalize with ASF/SF2, and these molecules not only regulate the variable splicing of precursor RNA but also participate in RNA transport (39). However, conversion between the two functions depends on the degree of phosphorylation, whereby excessive phosphorylation of ASF/SF2 participates in alternative RNA splicing (40), while a decrease in phosphorylation leads to interaction between ASF/SF2 and the TAP-P15 complex. This promotes the transport of mRNA to the outside of the nucleus (41). However, ALKBH5 knockout not only influenced ASF/SF2 expression but also altered subcellular localization of SRPK1 (a protein kinase related to ASF/SF2 phosphorylation) from the nucleus to the cytoplasm. This indirectly influenced the level of ASF/SF2 phosphorylation and therefore played a role in influencing mRNA nuclear transport (42). The above mechanisms are illustrated in Figure 1 and Table 2.
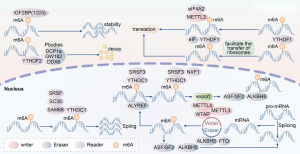
Table 2
| Classification of m6A regulators | Symbol of the regulators | Sub-cellular location | Biological function in the metabolism of RNAs | Reference |
|---|---|---|---|---|
| Readers | YTHDF2 | Cytoplasm | Mediating the intracellular RNA degradation by co-localizing with DCP1a, GW182, and DDX6, which are the component of RNA degradation related P-bodies | (23) |
| YTHDF1 | Cytoplasm | Accelerating the protein translation process by recruiting ribosomes and eIF3 to transcripts | (24,25) | |
| YTHDC1 | Nucleus | YTHDC1 can combine with SAM68, SC35, and SRSF, mediating the alternative splicing. Besides, the YTHDC1 can bind to the SRSF3 protein leading to the transportation of mRNA from nucleus to cytoplasm by binding NXF1 protein. YTHDC1 knock out witnessed no significant change in m6A/A ratios by LC-MS/MS, but in situ fluorescence hybridization reported increased m6A-methylated transcripts which is NXF1 independent. Over-expression of YTHDC1 decreased m6A-methylated mRNA transcripts in the nucleus but did not increase the number of m6A-methylated transcripts in the cytoplasm. YTHDC1 is also involved in intranuclear degradation of m6A-labeled mRNA | (26-30) | |
| Writer | METTL3 | Nucleus | METTL3 homolog Ime4 in Drosophilia is involved in alternating splicing of sex-lethal gene of drosophila, SXL. METTL3 can also interact with eIF4A2 to facilitate gene expression | (31,32) |
| WTAP, KIAA1429, and RBM15/RBM15B | Nucleus | There is evidence that splicing regulators such as Fl(2)D, Virilizer33, and Spenito (Nito) are homologous to the mammalian m6A methylation transferases. However, whether m6A modification is distributed near introns or exons close to the alternative splicing site is difficult to determine. | (33-35) | |
| Eraser | FTO | Nucleus | The deletion of FTO improves the mRNA binding capacity of SRSF2 resulting in an increase in the expression of the target exon. FTO is also involved in the RNA precursor splicing process | (36-38) |
| ALKBH5 | Nucleus | ALKBH5 co-locate with ASF/SF2, and these molecules not only regulate the variable splicing of precursor RNA | (39-42) |
YTHDF1/2, YTH domain family 1/2; YTHDC1, YTH domain containing 1; eIF, eukaryotic initiation factor; SAM68, Src-associated in mitosis 68KD; SRSF, serine/arginine-rich splicing factor; NXF1, nuclear RNA export factor 1; m6A, N6-adenosine methylation; LC-MS/MS, liquid chromatography coupled to tandem mass spectrometry; WTAP, Wilms tumor 1-associating protein; FTO, fat mass and obesity-associated gene; ALKBH5, AlkB homolog 5.
Impact of m6A modifications on signal transduction pathways
m6A can affect signal transduction pathways [such as the p53-mediated pathway, Notch signaling, nutrient sensing through the mammalian target of rapamycin complex 1 (mTORC1), and apoptosis] at the transcriptional level. For example, m6A can modify cyclin D1, which is involved in cell cycle regulation, to degrade its transcript, culminating in cell cycle stagnation at the G1/S checkpoint (43). However, the effect of m6A methylation on the cell cycle is controversial (44). Changes to m6A modifications can accelerate cell cycle transition from the G1 to the S phase by stabilizing CCNE1 mRNA (45), which is a regulatory subunit of cyclin-dependent kinase 2 (CDK2) that is central to the initiation of DNA replication at the G1/S checkpoint (46).
HF is a disease predominantly characterized by excessive apoptosis of cardiac myocytes, in which m6A has a crucial role. For example, in the mitochondria-dependent apoptosis pathway, BCL-2, which is located in the outer membrane of mitochondria and controls the outflow of cytochrome C from mitochondria, can bind to Bcl2 modifying factor (BMF), promote the release of cytochrome C, and promote apoptosis. YTHDF2, which recognizes the m6A methylation sequence, can bind to the BMF transcript and promote its degradation to reduce its protein expression, thus reducing apoptosis. Furthermore, in-vitro experiments showed that m6A methylation was enriched in the 3'-UTR region of the mTOR and PIK3C genes. Subsequent inhibition of m6A modification increased expression of mTOR and PIK3C. These changes may be caused by prolongation of the half-life of RNA transcripts resulting from decreased methylation of m6A (47). Therefore, these mechanisms suggest that m6A may be involved in the pathophysiological process of HF, and the methylation modifications of m6A and m6A regulatory molecules may serve as a new therapeutic target for HF (44).
m6A and lncRNA
As a new class of epigenetic regulatory factors, long noncoding RNAs (lncRNAs) are involved in the incidence and development of various diseases and have diagnostic value as biomarkers (48). The lncRNAs were mainly focused on its regulatory mechanisms in diseases, but how lncRNAs were regulated was less studied (49). Currently, an increasing number of studies has focused on the regulation of lncRNA by post-transcriptional modifications. Recent studies have reported that several lncRNAs, including MALAT1, MEG3, XIST, GAS5, and KCNK15-AS1, have undergone a widespread post-transcriptional modification of m6A (50). This suggests that m6A-modified lncRNAs play a substantial role in disease. Currently, there is no direct evidence that m6A-modified lncRNA plays an important role in any pathophysiological processes in HF. In 2021, Gong et al. proposed that lncRNA, ZFAS1, reduces the risk of atherosclerosis by promoting the expression of ADAM10/RAB22A, but ZFAS1 has m6A methylation sites that can be bound by METTL14 (51). Therefore, there is reason to believe that m6A methylation is involved in the development of atherosclerosis, a risk factor for HF, through the METTL14/ZFAS1/RAB22A pathway. Furthermore, studies have shown that ZFAS1 is involved in myocardial cell apoptosis induced by ischemia-reperfusion injury (52). However, the role of m6A methylation modifications in this process has not been thoroughly studied.
m6a modifications and HF
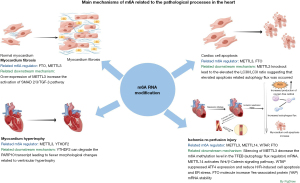
m6A RNA modification is involved in the regulation of environmental homeostasis in the cardiovascular system and the pathophysiological process of various cardiovascular diseases, regulating the stability of mRNA and changes in protein expression (53). Regulation of m6A modification of various homeostasis regulators is conducted at the post-transcriptional level, and this regulation mode responds rapidly to external signaling molecules and stimuli. As one of the most abundant RNA methylation modifications, there is increasing evidence that m6A is an important physiological mechanism in the incidence and development of HF (shown in Figure 2). The following paragraphs will elaborate on various pathophysiological mechanisms of m6A modification and its regulatory factors in HF (54).
m6a and myocardial fibrosis
Interstitial fibrosis is a marker of various heart diseases (55). Cardiac remodeling can initially maintain cardiac function based on molecular, cellular, and interstitial changes in the heart. However, if damaging stimuli remain, remodeling may cause a progressive and irreversible reduction in myocardium compliance (56). As an important component involved in myocardial fibrosis, the cardiomyocyte extracellular matrix (ECM) is not only a structural scaffold but also actively participates in various cellular signal-transduction activities and progressive reduction in cardiac compliance (57). Since cardiomyocytes are nonregenerative cells, when cardiomyocytes undergo apoptosis due to ischemia or stress, cardiac fibroblasts (CFs) are transformed into activated myofibroblasts. These myofibroblasts produce excessive ECM proteins to promote fibrosis to maintain the structural and functional integrity of the heart. However, excessive ECM causes pathological fibrotic remodeling in the ventricles, which affects the compliance of the ventricular wall and ultimately leads to congestive HF. Therefore, myocardial fibrosis is an important pathophysiological process in the progression of HF, and effective reversal of fibrosis can play a role in myocardial repair. The development of epigenetics has revealed that epigenetic modifications, such as DNA methylation, histone modification, and noncoding RNA regulation, are involved in the transcription of pro-fibrotic genes in cardiac fibrosis (58). However, the role of m6A modification in the process of myocardial fibrosis has not been thoroughly studied. Analysis of cardiac cells isolated from humans, mice, and pigs with HF showed that m6A levels in cardiac cells extracted from HF tissue were significantly increased compared with those of normal cardiac tissue. Expression of the m6A demethylation regulator FTO was also significantly decreased in HF tissues. Transfection of cells from the mouse myocardium with an AAV9 (aavFto) virus vector to stabilize the expression of FTO before and after the occurrence of simulated myocardial ischemia improved chronic cardiac function. Specifically, FTO-overexpressing mice had higher ejection fractions, improved ventricular wall movement, and increased systolic ratios. Meanwhile, quantitative analysis of ventricular fibrosis and angiogenesis showed that FTO overexpression effectively reduced myocardial fibrosis caused by ischemia, delaying myocardial remodeling and improving cardiac function. Simultaneously, the in vitro FTO knockout myocardial cells exhibited significantly more arrhythmia events (59). The severe decrease in the ejection fraction and the severe increase in ventricular dilatation in FTO knockout mice promoted the progression of HF (60). Therefore, dysregulated m6A modification of mRNA is a marker of HF, while FTO demethylase is closely related to improved cardiac function. Therefore, FTO is a potential therapeutic target of HF. Conversely, overexpressing METTTL3 (m6A methylation writer) can increase m6A levels. Activation of the Sma and mad related protein 2/3 (SMAD2/3), a key signaling molecule that promotes the transforming growth factor-β1 (TGF-β1) pathway, results in altered transcription levels of many cardiac fibrosis-related genes and mediates the synthesis of large amounts of collagen (61).
m6A and ventricular hypertrophy
Another major manifestation of HF is ventricular hypertrophy, a pathophysiological presentation associated with excessive ventricular afterload. Compensatory fibro-thickening occurs, and new myocardial fibers are produced to overcome the increased afterload. These changes can be observed macroscopically as centripetal ventricular remodeling, with gradual increases in ventricular wall thickness and decreases in volume of the ventricular lumen, accompanied by an increase in left ventricular mass. In the short term, these changes are compensatory, maintaining sufficient myocardial systolic force to match cardiac output with increased afterload (62). Ventricular hypertrophy is caused by a change in expression of specific genes in cardiac myocytes that leads to increased production of protein molecules associated with cardiac hypertrophy. The activation of transcription factors via signal transduction pathways related to cardiac hypertrophy have been explored, but few studies have focused on the effects of post-transcriptional modification on cardiac hypertrophy (63). In 2020, whole-genome sequencing of m6A was used to analyze methylated transcripts from HF and non-HF tissues and of HF and the ventricular-hypertrophic myocardium. The HF group had significantly increased levels of m6A modifications. Gene Ontology (GO) cellular component enrichment analysis of m6A transcripts in HF tissue were significantly enriched in histone methylation function. In addition, 37% of the m6A-modified transcripts overlapped between HF cardiac tissue and ventricular hypertrophy tissue. GO enrichment analysis of these overlapping transcripts revealed that most of the transcripts were related to the regulation of gene expression. This evidence suggested that m6A modification has a crucial bridging role in the regulation of gene transcription and post-transcriptional modification in the process of HF and ventricular hypertrophy (64). Studies have proven that m6A and METTL3 play key roles in the development of cardiac hypertrophy, but the results of these studies are inconsistent. By m6A genome sequencing and m6A immunoprecipitation, Dorn et al. found that m6A levels were significantly upregulated among all mRNA levels in hypertrophic ventricular muscle. Furthermore, m6A was significantly enriched in protein kinase-related transcripts. Meanwhile, METTL3 overexpression in vivo and in vitro resulted in the lengthening and widening of cardiac cells. An increased heart mass-to-body weight ratio was observed in in-vivo experiments modeling cardiac hypertrophy. However, no histopathological changes were observed. Concurrently, cardiac ultrasound showed that cardiac function was unaffected in experimental animals after METTL3 overexpression (15). Elevated PARP10 expression in the hypertrophic myocardium was first identified using a trans-aortic constriction (TAC)-induced ventricular hypertrophy model. Meanwhile, TAC-treated myocardial tissue in PARP10-knockout mice had fewer morphological changes related to ventricular hypertrophy, and a reduced heart mass, compared with control mice. These mice also had a lower reduction in heart function. m6A sequencing analysis of PARP10 in myocardial cells of mice with ventricular hyperplasia showed that m6A modification of PARP10 decreased significantly, and this decrease was positively correlated with CHAPIR. In addition, after CHAPIR expression was knocked out, the degree of binding between METTL3 and PARP10 increased. Subsequent analysis showed that the increase in m6A modification of PARP10 was related to increased transcript degradation, and the transcription degradation process was YTHDF2-dependent. It was therefore concluded that m6A modification is involved in the post-transcriptional regulation of some molecules related to ventricular hypertrophy (65).
m6A and myocardial cell apoptosis
HF is the final stage of all cardiovascular diseases. Although there are many pathophysiological mechanisms of HF, cardiomyocyte apoptosis is the most direct cause and is characteristic of HF. METTL3 is the predominant factor involved in abnormal m6A modification. A significant increase in METTL3 was found in myocardial cells treated with hypoxia. By exploring the ratio of LC3II to LC3I, it was found that light chain 3 (LC3)II/LC3I increased significantly if METTL3 expression was knocked out. The increased ratio may be related to increased LC3 lipidation levels caused by increased autophagy or lysosomal and autophagosomal degradation. METTL3-silenced H9c2 cells were treated with the lysosomal inhibitor bafilomycin A1 (BafA) and, compared with the control group, the LC3II/LC3I ratio was significantly increased after METTL3 knockout, suggesting that elevated autophagy flux occurred (66). In addition, transfection of neonatal mouse ventricular myocytes with a MRFP-GFP-LC3 vector demonstrated that when METTL3 expression was knocked out, there were more autophagic lysosomes and fewer autophagosomes, which indirectly supported that METTL3 could inhibit the autophagy flux, while cell apoptosis was often associated with inhibition of autophagy. The degree of hypoxia-induced apoptosis of cardiomyocytes also decreased after METTL3 deletion. Conversely, the specific mechanisms involving METTL3 in apoptosis are as follows. METTL3 can decrease transcription stability and expression by increasing transcription factor EB (TFEB) RNA methylation. TFEB, as a transcription factor in the coordinated lysosomal expression and regulation (CLEAR) network, regulates the expression of various autophagy-related molecules and mediates lysosomal pathways, which play an important role in the integrity of autophagy (67,68). Therefore, m6A and its regulatory factors have a regulatory effect on autophagy-mediated apoptosis (69). In the regulation of m6A on noncoding RNAs associated with apoptosis, FTO (another regulator of m6A modifications) increased myosin heavy chain associated RNA transcript (MHRT) expression and reduced myosin heavy chain-associated RNA transcripts in hypoxia-treated cardiomyocytes. MHRT can inhibit apoptosis of myocardial cells (70). Simultaneously, increased methylation and decreased expression of MHRT were observed in a mouse model of HF. m6A modification often leads to degradation of transcription stability and decreased expression of MHRT. The main function of FTO is demethylation, which reduces the degree of methylation of m6A. Therefore, increased cardiomyocyte apoptosis, caused by decreased expression of FTO, may be related to decreased expression of MHRT, which is indirectly caused by increased methylation of MHRT (71). In addition, the main function of MHRT protein depends on the downstream molecule Brg1, which is a protein related to histone modification. Therefore, m6A modification is related to other epigenetic modifications, such as chromosomal modifications, to affect cell apoptosis and can be considered a new mechanism (72). Besides autophagy and the MHRT pathway, a recent study showed that FTO in skeletal muscles can mediate the demethylation of GADD45B mRNA by m6A and increase the stability of the GADD45B transcript, which is an activator of the p38 MAPK pathway (73). However, activating the MAPK signaling pathway can reduce the area of myocardial infarction in ischemia by reducing myocardial cell apoptosis (74). Thus, FTO may regulate cardiac cell apoptosis through the MAPK signaling pathway; however, this hypothesis has not been validated in cultured cardiac cells.
m6A and myocardial ischemia reperfusion injury (IRI)
Acute myocardial infarction (AMI) is a leading cause of HF. Reperfusion therapy is an important aspect in improving the prognosis of patients with HF. After performing reperfusion, a secondary injury can be caused due to the production of radical oxygen species (ROS) and subsequent activation of inflammation. Thus, IRI can eliminate the clinical benefits of reperfusion therapy. Further investigation of the mechanism of IRI is therefore warranted. Recent studies have found a connection between m6A modification and IRI. Song et al. reported that under hypoxia and reoxygenation, H9c2 cells and the NMVCs cell line presented significantly elevated levels of m6A modification in dot blot analysis. The same result was obtained following in-vivo simulation of IRI in mice (69). This alternation of m6A modification was believed to be the result of alternation of m6A modification regulator molecules. Subsequent quantification analysis of m6A regulators at the gene level revealed that only METTL3 exhibited a statistically significant elevation in the ischemic/reperfusion heart. Thus, exploration of the function of METTL3 in IRI process is continuing. As mentioned above, METTL3 is negatively related to the autophagy flux, and reducing autophagy can lead to apoptosis (75). Silencing METTL3 decreased the level of m6A in TFEB mRNA in vitro. The TFEB gene is essential for regulating autophagy flux and lysosomal biogenesis (68). Thus, METTL3 reduces autophagy in IRI cardiac cells by reducing the expression of TFEB through increasing m6A modification in the transcript and decreasing transcript stability. Moreover, in 2021, METTL14 was also found to be associated with the protection of cardiomyocytes from infarction/reperfusion (I/R) via activation of the Wnt/β-catenin signaling pathway in a m6A-dependent manner (76). As another class of m6A regulator, WTAP suppressed ATF4 expression by increasing the m6A level of the ATF4 transcript, reducing hypoxia/reperfusion (H/R)-induced cell apoptosis and endoplasmic reticulum (ER) stress in AC16 cells (77). Furthermore, the FTO molecule can protect cardiomyocytes via increasing Yes-associated protein (YAP) mRNA stability (78); YAP is associated with proliferation and oncogenic activity (79). Collectively, this evidence indicates that alternation of m6A and m6A regulator could potentially be used to curb the damage caused by IRI.
The role of m6A in HF-related immune cell infiltration
Except for the mechanisms mentioned above (summarized in Table 3). Immune cell infiltration also plays a key role in the incidence of HF. When cardiomyocytes become necrotic under conditions of ischemia and hypoxia, their cell fragments will act as endogenous immunogens to trigger the activation of endogenous immune responses. For example, to induce immune cell infiltration, neutrophils access the myocardium and secrete matrix metalloproteinases (MMPs) to help remove cell debris and secrete IL-6 and other macrophage chemoattractants. In addition, when the autoantigen is recognized by antigen-presenting cells, it activates CD4+ T cells and induces their differentiation into specific subtypes. Activation of macrophages residing in the ECM leads to the release of the cytokine tumor necrosis factor (TNF)-alpha, which is involved in cardiac remodeling (80,81). METTL3 directly regulates the signal transducer and activator of transcription 1 (STAT1) at the post-transcriptional level, polarizing macrophages towards the M1 subtype (82). However, in the pathophysiological process of ventricular remodeling and ventricular hypertrophy, M1-polarized macrophages aggravate myocardial injury by producing many inflammatory factors. Studying the regulatory mechanism of m6A modifications in immune cell infiltration can therefore provide new therapeutic targets for the treatment of HF (83). However, there is no direct evidence to show the relationship between m6A and immune infiltration in the HF process. Thus, future research should focus on the influence of m6A on immune cell infiltration in the pathophysiological process of HF.
Table 3
| Patho-physiology process of HF | Evidence of m6A participating in HF progression | Reference |
|---|---|---|
| Myocardium fibrosis | Interstitial fibrosis result in reduction in myocardium compliance involving various types of molecular, cellular, and interstitial changes | (55-61) |
| Overexpression of FTO improve cardiac function by reducing the collagen deposition | ||
| Over-expression of METTL3 increase the activation of SMAD2/3/TGF-β pathway leading to the accumulation of collagen | ||
| Ventricular hypertrophy | Ventricular hypertrophy is a temporary compensatory mechanism for the increasing after load, which is manifested by ventricular wall thickness, decreases in volume of the ventricular lumen and increase in left ventricular mass | (15,62-65) |
| Hypertrophic failing hearts witnessed higher level of m6A modification verified by m6A sequencing. The failing hearts have overlapped m6A modified transcripts with those hypertrophy heart tissue, which were related to the regulation of gene expression | ||
| METTL3 overexpression in in vivo and in vitro experiments resulted in the lengthening and widening of cardiac cells | ||
| YTHDF2 can degrade the PARP10 transcript. The PARP10 showed elevation in the failing myocardium and knocking down of PARP10 leads to fewer morphological changes related to ventricular hypertrophy and reduced heart mass | ||
| Cardiac cell apoptosis | METTL3 showed correlation with the autophagy process, since METTL3 knockout led to the elevated the LC3II/LC3I ratio suggesting that elevated autophagy flux was occurred, while cell apoptosis was often associated with inhibition of autophagy | (66-74) |
| m6A eraser FTO were shown to increase MHRT expression and reduce myosin heavy chain-associated RNA transcripts in hypoxia-treated cardiomyocytes. MHRT can inhibit apoptosis of myocardial cells | ||
| m6A modification is related to chromosomal modifications. FTO can mediate the demethylation of GADD45B mRNA and increase the stability of the GADD45B transcript, which is an activator of the p38 MAPK pathway mediating reduction of the area of myocardial infarction | ||
| Ischemic reperfusion injury | The myocardium undergone ischemic reperfusion injury showed elevated level of m6A modification both in cell-line and in vivo studies | (75,76,78-80) |
| METTL3 increase the m6A methylation level in the TFEB mRNA and subsequently decrease autophagy flux and lysosomal biogenesis in hypoxia/reoxygenation-treated cardiomyocytes | ||
| METTL14 protect cardiomyocytes from I/R via the activation of Wnt/β-Catenin signaling pathway in m6A dependent manner | ||
| WTAP suppressed H/R-induced cell apoptosis and ER stress in AC16 cells by increasing the m6A level of ATF4 transcription script | ||
| FTO molecule can protect the cardiomyocyte via increasing YAP mRNA stability, which is related to the proliferation and oncogenic activity |
m6A, N6-adenosine methylation; HF, heart failure; FTO, fat mass and obesity-associated gene; METTL3, methyltransferase-like 3; SMAD2/3, Sma and mad related protein 2/3; TGF-β, transforming growth factor β; YTHDF2, YTH domain family 2; LC3, light chain 3; MHRT, myosin heavy chain associated RNA transcript; GADD45B, growth arrest and DNA damage inducible beta; TFEB, transcription factor EB; METTL14, methyltransferase-like 14; I/R, infarction/reperfusion; WTAP, Wilms tumor 1-associating protein; H/R, hypoxia/reperfusion; ER, endoplasmic reticulum; YAP, Yes-associated protein.
The role of m6A regulatory factors in the clinical diagnosis and treatment of HF
METTL3
Full-length METTL3 comprises 580 amino acids and consists of a zinc finger domain (ZFD) and a methyltransferase domain, both of which are required for enzyme activity (84). Recently, METTL3 was suggested to be involved in the occurrence and development of HF. Kmietczyk et al. explored the association between m6A modification and HF and found that m6A levels increased in failing human cardiomyocytes. Further animal experiments showed that METTL3 knockout significantly increased the expression of the HF markers, atrial natriuretic peptide (ANP) precursors A and B (85). However, increased levels of ANP are associated with a reduction in ejection fraction, long-term improvement in HF, and relief of ventricular remodeling (86). Therefore, METTTL3 could be used as a new therapeutic target to improve the prognosis of HF. Recent studies by Zhang et al. suggested that m6A modification also plays a key role in HF with retention of the ejection fraction. Sixteen heart failure with preserved ejection fraction (HFpEF) patients were selected as the study subjects and were compared with 24 healthy controls during the same period. Real-time PCR (RT-PCR) was used to evaluate the m6A regulatory factor in the peripheral blood of HFpEF patients and healthy controls. The patients with HFpEF exhibited higher expression of METTL3 in peripheral blood compared with healthy controls, thus the expression of METTL3 was different in different tissue sources. This study was the first to assess changes in METTL3 in peripheral blood of HFpEF patients, providing a basis for further research on the diagnostic potential of METTL3 (87).
FTO
FTO is instrumental in primary diseases related to HF, such as hypertrophic cardiomyopathy, ventricular septal defect (88), arrhythmia (89), coronary heart disease, and ischemic cardiomyopathy (90). Mathiyalagan et al. quantified m6A levels in left ventricular myocardial tissue from human patients with HF (ischemic and non-ischemic) and in pigs and mice with HF following myocardial infarction, compared with control groups without HF. The experiment results shows that the levels of total RNA and polyA+ RNA m6A in human, pig, and mouse left ventricular tissues were significantly higher compared with those of the control group. Expression of the regulatory proteins for m6A modification in human and mouse hearts, including METTL3, METTL4, METTL14, WTAP, FTO, and ALKBH5, were also investigated. RNA levels and protein expression levels of FTO were significantly reduced compared with their respective control groups. Four hours after myocardial infarction, the level of FTO decreased in the ischemic myocardial tissue of mice and this decrease in FTO mRNA and protein levels was consistent with the increase of m6A in mice at 1 and 4 weeks after myocardial infarction. Furthermore, changes in METTL3, METTL4, METTL14, WTAP, and ALKBH5 could not completely explain the abnormal continuous increase in m6A in failing hearts. These data suggest that ischemia-induced FTO loss may be an important molecular marker that explains the increase in m6A in failing hearts in humans and mice. Furthermore, FTO overexpression improved cardiac function in ischemia-induced HF mouse models by reducing myocardial fibrosis and angiogenesis (91). Berulava et al. constructed a mouse model of myocardial hypertrophy by aortic coarctation and found that m6A levels gradually increased and FTO expression gradually decreased in the process of cardiac hypertrophy developing into HF. Compared with the control group, knockout of the FTO gene in mice resulted in a more severe decrease in cardiac ejection fraction and a higher degree of cardiac dilation. Further experiments found that hypermethylated and hypomethylated transcripts could be detected in HF mice, suggesting that over-methylation is involved in HF (60). These cardiac events were related to the calcium (Ca2+) dynamics in cardiac cells. Sarcoplasmic reticulum calcium ATPase (SERCA) 2a is a major subtype of SERCA in cardiomyocytes and has a vital role in the regulation of calcium equilibrium (92). Modulation of SERCA2a expression with gene transfer improved contractility and energy supply and utilization (93). By comparing RNA methylation sequencing patterns, it was shown that m6A modulation of SERCA2a in ischemic cardiomyocytes was demethylated by FTO, which prevented the degradation of SERCA2a mRNA and increased the stability and protein levels of SERCA2a (41). Altogether, these experiments confirmed that the decrease in FTO levels was closely related to the increase in m6A levels and the incidence and development of HF. This further confirmed the potential application of FTO in the treatment of HF. Targeting regulation of FTO or m6A methylation may be a promising therapeutic strategy for HF. In addition, heart transplantation is a relatively common treatment for end-stage HF. Some studies have used matched samples from 370 heart transplantation operations to show that the polymorphism of the RS17817449 site of the FTO gene in donor hearts is related to immune rejection after transplantation, and donors with a TT allele are more likely to promote immune rejection after transplantation compared with the individuals with at least one G allele (94).
Other findings related to the value of m6A in the clinical diagnosis and treatment of HF
m6A, as a prevalent post-transcriptional epigenetic modification of mRNA in mammals (95), has been investigated in various types of cardiovascular disease. For example, the Ischemic Heart Disease Epitranscriptomics and Biomarkers (IHD-EPITRAN) study investigated the potential of m6A modifications of RNA as a biomarker to predict the occurrence of ischemic heart disease (96). Furthermore, exploration of m6A-single nucleotide polymorphisms (m6A-SNPs) in a genome-wide association study (GWAS) involving 188,578 subjects revealed that the m6A-SNP rs6859 in the 3’-UTR of PVRL2 was associated with multiple types of molecules related to lipid metabolism, including high-density lipoprotein cholesterol, low-density lipoprotein (LDL) cholesterol, total cholesterol, and triglycerides (97). LDL plays a causal role in coronary artery disease (98). Meanwhile, m6A also played a significant role in the pathogenesis of hypertension. In 2019, researchers investigated the association between m6A-SNPs and blood pressure via a large-scale GWAS and found that there were 1,236 m6A-SNPs that were nominally associated with blood pressure, with 10% of them also associated with coronary artery disease or stroke (99). These results proved that the modifications of m6A served as a marker of diverse types of cardiovascular disease, and the diseases mentioned above have always served as an etiology of HF. For example, hypertension leads to chronic pressure overload, resulting in left ventricular hypertrophy and subsequent dilation disorders. Persistent hypertrophy and dilation disorders can cause HF with dilated cardiomyopathy (100). Regarding the potential of m6A as a therapeutic target, Kumari et al. found that ALKBH5 facilitated improvement of angiogenesis after acute ischemic stress by decreasing the m6A level of SPHK1 transcripts; SPHK1 regulated the phosphorylation of endothelial nitric oxide synthase (eNOS) and PI3K/AKT, two important signaling pathways for endothelium angiogenesis (101). Thus, ALKBH5 can serve as a potential therapeutic target for HF.
Future perspectives
As a terminal state of diverse types of cardiovascular diseases, more research should be dedicated to exploring the molecular aspects underlying the pathophysiological mechanism of HF to define suitable therapeutic targets. As one pathological process of HF, cardiomyopathic hypertrophy serves as an adaptive response to volume overload. However, with the persistence of myocardial hypertrophic remodeling, the compliance of the myocardium decreases and HF ultimately occurs. A recent study using the methylated RNA immunoprecipitation sequencing (meRIP-seq) technique revealed that the m6A level in 16 mRNAs associated with hypertrophy was significantly increased compared with the normal myocardium, and alternation of the m6A RNA regulator, METTL3, plays an important role in the reversal of cardiomyocyte hypertrophy in vitro and in vivo (102). Thus, targeting m6A regulators may be a promising therapy for HF. Recent research efforts have investigated the alteration of the landscape of m6A modification, the expression of m6A regulators, and their effect on protein expression levels (87). However, further research on m6A modifications in HF is worthwhile because there is currently no direct evidence to prove the relationship between m6A modification and several important pathophysiological mechanisms related to HF. For instance, the correlation between immune cell infiltration in failing hearts and m6A patterns has not been investigated in detail. Bioinformatics techniques may facilitate in making the connection between myocardium immune cell infiltration and m6A modification (103). Furthermore, the interplay between m6A and other epigenetic modifications in the progression of HF has not been researched. Exploring these mechanisms will improve understanding of the mechanisms of HF and aid in the search for novel therapeutic targets. Moreover, more focus is needed on the interaction between the m6A regulators and targeted transcript(s), and immune precipitation experiments between the target transcript(s) and m6A regulators may help prove that m6A regulators do have an action on the targeted transcript.
Currently, there is considerable research focusing on the involvement of m6A in the risk factors of HF like atherosclerosis, hypertension, and diabetes (104). However, research on the role of m6A modification in diabetic heart diseases and HF is rare. In diabetic patients, the cardiac lesion predominantly presents as diastolic dysfunction, which is an initial stage of HF (105). Thus, future investigations should focus on subjects with diabetic myopathy and atrial fibrillation, respectively, and additional work should be devoted to the association between HF risk factor and the m6A modification level in peripheral circulation cellular components, with the aim of improving application of the research results to clinical settings.
Acknowledgments
Thanks for our country!
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://cdt.amegroups.com/article/view/10.21037/cdt-22-277/rc
Peer Review File: Available at https://cdt.amegroups.com/article/view/10.21037/cdt-22-277/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://cdt.amegroups.com/article/view/10.21037/cdt-22-277/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ponikowski P, Voors AA, Anker SD, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC)Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J 2016;37:2129-200. Erratum in: Eur Heart J 2018 Mar 7;39(10):860. [Crossref] [PubMed]
- Tsao CW, Aday AW, Almarzooq ZI, et al. Heart Disease and Stroke Statistics-2022 Update: A Report From the American Heart Association. Circulation 2022;145:e153-639. [Crossref] [PubMed]
- Dunlay SM, Roger VL, Redfield MM. Epidemiology of heart failure with preserved ejection fraction. Nat Rev Cardiol 2017;14:591-602. [Crossref] [PubMed]
- Di Salvo TG. Epigenetic Regulation in Heart Failure: Part I RNA. Cardiol Rev 2015;23:213-28. [Crossref] [PubMed]
- Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017;169:1187-200. [Crossref] [PubMed]
- Zhang DD, Shen Y, Hong K. Research update of RNA N6-methyladenosine modification in cardiovascular diseases. Zhonghua Xin Xue Guan Bing Za Zhi 2021;49:401-4. [PubMed]
- Zhao K, Yang CX, Li P, et al. Epigenetic role of N6-methyladenosine (m6A) RNA methylation in the cardiovascular system. J Zhejiang Univ Sci B 2020;21:509-23. [Crossref] [PubMed]
- Martinez SR, Gay MS, Zhang L. Epigenetic mechanisms in heart development and disease. Drug Discov Today 2015;20:799-811. [Crossref] [PubMed]
- Wu Y, Zhan S, Xu Y, et al. RNA modifications in cardiovascular diseases, the potential therapeutic targets. Life Sci 2021;278:119565. [Crossref] [PubMed]
- Longenecker JZ, Gilbert CJ, Golubeva VA, et al. Epitranscriptomics in the Heart: a Focus on m6A. Curr Heart Fail Rep 2020;17:205-12. [Crossref] [PubMed]
- Dong L, Cao Y, Hou Y, et al. N6 -methyladenosine RNA methylation: A novel regulator of the development and function of immune cells. J Cell Physiol 2022;237:329-45. [Crossref] [PubMed]
- Brocard M, Ruggieri A, Locker N. m6A RNA methylation, a new hallmark in virus-host interactions. J Gen Virol 2017;98:2207-14. [Crossref] [PubMed]
- Huisman B, Manske G, Carney S, et al. Functional Dissection of the m6A RNA Modification. Trends Biochem Sci 2017;42:85-6. [Crossref] [PubMed]
- Wang K, Li Y, Qiang T, et al. Role of epigenetic regulation in myocardial ischemia/reperfusion injury. Pharmacol Res 2021;170:105743. [Crossref] [PubMed]
- Dorn LE, Lasman L, Chen J, et al. The N6-Methyladenosine mRNA Methylase METTL3 Controls Cardiac Homeostasis and Hypertrophy. Circulation 2019;139:533-45. [Crossref] [PubMed]
- Papait R, Corrado N, Rusconi F, et al. It's Time for An Epigenomics Roadmap of Heart Failure. Curr Genomics 2015;16:237-44. [Crossref] [PubMed]
- Fu Y, Dominissini D, Rechavi G, et al. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet 2014;15:293-306. [Crossref] [PubMed]
- Kierzek E, Kierzek R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res 2003;31:4472-80. [Crossref] [PubMed]
- Zhang X, Lu N, Wang L, et al. Recent advances of m6A methylation modification in esophageal squamous cell carcinoma. Cancer Cell Int 2021;21:421. [Crossref] [PubMed]
- Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 2007;318:1469-72. [Crossref] [PubMed]
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011;7:885-7. [Crossref] [PubMed]
- Fang Z, Hu Y, Hu J, et al. The crucial roles of N6-methyladenosine (m6A) modification in the carcinogenesis and progression of colorectal cancer. Cell Biosci 2021;11:72. [Crossref] [PubMed]
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117-20. [Crossref] [PubMed]
- Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015;161:1388-99. [Crossref] [PubMed]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010;11:113-27. [Crossref] [PubMed]
- Hartmann AM, Nayler O, Schwaiger FW, et al. The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol Biol Cell 1999;10:3909-26. [Crossref] [PubMed]
- Imai Y, Matsuo N, Ogawa S, et al. Cloning of a gene, YT521, for a novel RNA splicing-related protein induced by hypoxia/reoxygenation. Brain Res Mol Brain Res 1998;53:33-40. [Crossref] [PubMed]
- Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 2019;20:608-24. [Crossref] [PubMed]
- Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 2017;6:e31311. [Crossref] [PubMed]
- Lubas M, Christensen MS, Kristiansen MS, et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell 2011;43:624-37. [Crossref] [PubMed]
- Ye F, Wang X, Tu S, et al. The effects of NCBP3 on METTL3-mediated m6A RNA methylation to enhance translation process in hypoxic cardiomyocytes. J Cell Mol Med 2021;25:8920-8. [Crossref] [PubMed]
- Lence T, Akhtar J, Bayer M, et al. m6A modulates neuronal functions and sex determination in Drosophila. Nature 2016;540:242-7. [Crossref] [PubMed]
- Hilfiker A, Amrein H, Dübendorfer A, et al. The gene virilizer is required for female-specific splicing controlled by Sxl, the master gene for sexual development in Drosophila. Development 1995;121:4017-26. [Crossref] [PubMed]
- Granadino B, Campuzano S, Sánchez L. The Drosophila melanogaster fl(2)d gene is needed for the female-specific splicing of Sex-lethal RNA. EMBO J 1990;9:2597-602. [Crossref] [PubMed]
- Xie S, Chen W, Chen K, et al. Emerging roles of RNA methylation in gastrointestinal cancers. Cancer Cell Int 2020;20:585. [Crossref] [PubMed]
- Zhao X, Yang Y, Sun BF, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 2014;24:1403-19. [Crossref] [PubMed]
- Bartosovic M, Molares HC, Gregorova P, et al. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res 2017;45:11356-70. [Crossref] [PubMed]
- Lan Q, Liu PY, Haase J, et al. The Critical Role of RNA m6A Methylation in Cancer. Cancer Res 2019;79:1285-92. [Crossref] [PubMed]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 2005;122:365-78. [Crossref] [PubMed]
- Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell 2004;13:201-12. [Crossref] [PubMed]
- Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell 2008;30:179-89. [Crossref] [PubMed]
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013;49:18-29. [Crossref] [PubMed]
- Hirayama M, Wei FY, Chujo T, et al. FTO Demethylates Cyclin D1 mRNA and Controls Cell-Cycle Progression. Cell Rep 2020;31:107464. [Crossref] [PubMed]
- Tan F, Zhao M, Xiong F, et al. N6-methyladenosine-dependent signalling in cancer progression and insights into cancer therapies. J Exp Clin Cancer Res 2021;40:146. [Crossref] [PubMed]
- Zhu W. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J Cell Mol Med 2020;24:3521-33. [Crossref] [PubMed]
- Caruso JA, Duong MT, Carey JPW, et al. Low-Molecular-Weight Cyclin E in Human Cancer: Cellular Consequences and Opportunities for Targeted Therapies. Cancer Res 2018;78:5481-91. [Crossref] [PubMed]
- Zhao Q, Zhao Y, Hu W, et al. m6A RNA modification modulates PI3K/Akt/mTOR signal pathway in Gastrointestinal Cancer. Theranostics 2020;10:9528-43. [Crossref] [PubMed]
- Aftabi Y, Ansarin K, Shanehbandi D, et al. Long non-coding RNAs as potential biomarkers in the prognosis and diagnosis of lung cancer: A review and target analysis. IUBMB Life 2021;73:307-27. [Crossref] [PubMed]
- Torsin LI, Petrescu GED, Sabo AA, et al. Editing and Chemical Modifications on Non-Coding RNAs in Cancer: A New Tale with Clinical Significance. Int J Mol Sci 2021;22:581. [Crossref] [PubMed]
- Cayir A. Environmental exposures and RNA N6-Methyladenosine modified long Non-Coding RNAs. Crit Rev Toxicol 2020;50:641-9. [Crossref] [PubMed]
- Gong C, Fan Y, Liu J. METTL14 mediated m6A modification to LncRNA ZFAS1/ RAB22A: A novel therapeutic target for atherosclerosis. Int J Cardiol 2021;328:177. [Crossref] [PubMed]
- Huang P, Yang D, Yu L, et al. Downregulation of lncRNA ZFAS1 protects H9c2 cardiomyocytes from ischemia/reperfusion-induced apoptosis via the miR-590-3p/NF-κB signaling pathway. Mol Med Rep 2020;22:2300-6. [Crossref] [PubMed]
- Devaux Y, Nossent AY. EU-CardioRNA COST Action CA17129. A role for m6A RNA methylation in heart failure development? Eur J Heart Fail 2020;22:67-9. [Crossref] [PubMed]
- Kumari R, Ranjan P, Suleiman ZG, et al. mRNA modifications in cardiovascular biology and disease: with a focus on m6A modification. Cardiovasc Res 2022;118:1680-92. [Crossref] [PubMed]
- Hu F, Li M, Han F, et al. Role of TRPM7 in cardiac fibrosis: A potential therapeutic target Exp Ther Med 2021;21:173. (Review). [Crossref] [PubMed]
- Gubert F, da Silva JS, Vasques JF, et al. Mesenchymal Stem Cells Therapies on Fibrotic Heart Diseases. Int J Mol Sci 2021;22:7447. [Crossref] [PubMed]
- Loffredo FS, Nikolova AP, Pancoast JR, et al. Heart failure with preserved ejection fraction: molecular pathways of the aging myocardium. Circ Res 2014;115:97-107. [Crossref] [PubMed]
- Li X, Yang Y, Chen S, et al. Epigenetics-based therapeutics for myocardial fibrosis. Life Sci 2021;271:119186. [Crossref] [PubMed]
- Mathiyalagan P, Adamiak M, Mayourian J, et al. FTO-Dependent N6-Methyladenosine Regulates Cardiac Function During Remodeling and Repair. Circulation 2019;139:518-32. [Crossref] [PubMed]
- Berulava T, Buchholz E, Elerdashvili V, et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail 2020;22:54-66. [Crossref] [PubMed]
- Li T, Zhuang Y, Yang W, et al. Silencing of METTL3 attenuates cardiac fibrosis induced by myocardial infarction via inhibiting the activation of cardiac fibroblasts. FASEB J 2021;35:e21162. [Crossref] [PubMed]
- Abecasis J, Gomes Pinto D, Ramos S, et al. Left Ventricular Remodeling in Degenerative Aortic Valve Stenosis. Curr Probl Cardiol 2021;46:100801. [Crossref] [PubMed]
- Akazawa H, Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res 2003;92:1079-88. [Crossref] [PubMed]
- Hinger SA, Wei J, Dorn LE, et al. Remodeling of the m6A landscape in the heart reveals few conserved post-transcriptional events underlying cardiomyocyte hypertrophy. J Mol Cell Cardiol 2021;151:46-55. [Crossref] [PubMed]
- Gao XQ, Zhang YH, Liu F, et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N6-methyladenosine methylation of Parp10 mRNA. Nat Cell Biol 2020;22:1319-31. [Crossref] [PubMed]
- Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012;8:445-544. [Crossref] [PubMed]
- Chua JP, Reddy SL, Merry DE, et al. Transcriptional activation of TFEB/ZKSCAN3 target genes underlies enhanced autophagy in spinobulbar muscular atrophy. Hum Mol Genet 2014;23:1376-86. [Crossref] [PubMed]
- Zhao E, Czaja MJ. Transcription factor EB: a central regulator of both the autophagosome and lysosome. Hepatology 2012;55:1632-4. [Crossref] [PubMed]
- Song H, Feng X, Zhang H, et al. METTL3 and ALKBH5 oppositely regulate m6A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy 2019;15:1419-37. [Crossref] [PubMed]
- Zhang L, Wu YJ, Zhang SL. Circulating lncRNA MHRT predicts survival of patients with chronic heart failure. J Geriatr Cardiol 2019;16:818-21. [PubMed]
- Shen W, Li H, Su H, et al. FTO overexpression inhibits apoptosis of hypoxia/reoxygenation-treated myocardial cells by regulating m6A modification of Mhrt. Mol Cell Biochem 2021;476:2171-9. [Crossref] [PubMed]
- Shen S, Jiang H, Bei Y, et al. Long Non-Coding RNAs in Cardiac Remodeling. Cell Physiol Biochem 2017;41:1830-7. [Crossref] [PubMed]
- Deng K, Fan Y, Liang Y, et al. FTO-mediated demethylation of GADD45B promotes myogenesis through the activation of p38 MAPK pathway. Mol Ther Nucleic Acids 2021;26:34-48. [Crossref] [PubMed]
- Ren J, Zhang S, Kovacs A, et al. Role of p38alpha MAPK in cardiac apoptosis and remodeling after myocardial infarction. J Mol Cell Cardiol 2005;38:617-23. [Crossref] [PubMed]
- Song H, Pu J, Wang L, et al. ATG16L1 phosphorylation is oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein phosphatase 1 which determines the fate of cardiomyocytes during hypoxia/reoxygenation. Autophagy 2015;11:1308-25. [Crossref] [PubMed]
- Pang P, Qu Z, Yu S, et al. Mettl14 Attenuates Cardiac Ischemia/Reperfusion Injury by Regulating Wnt1/β-Catenin Signaling Pathway. Front Cell Dev Biol 2021;9:762853. [Crossref] [PubMed]
- Wang J, Zhang J, Ma Y, et al. WTAP promotes myocardial ischemia/reperfusion injury by increasing endoplasmic reticulum stress via regulating m6A modification of ATF4 mRNA. Aging (Albany NY) 2021;13:11135-49. [Crossref] [PubMed]
- Ke WL, Huang ZW, Peng CL, et al. m6A demethylase FTO regulates the apoptosis and inflammation of cardiomyocytes via YAP1 in ischemia-reperfusion injury. Bioengineered 2022;13:5443-52. [Crossref] [PubMed]
- Yu H, Tang H, Deng C, et al. RRM2 Improves Cardiomyocyte Proliferation after Myocardial Ischemia Reperfusion Injury through the Hippo-YAP Pathway. Dis Markers 2021;2021:5089872. [Crossref] [PubMed]
- Rai A, Narisawa M, Li P, et al. Adaptive immune disorders in hypertension and heart failure: focusing on T-cell subset activation and clinical implications. J Hypertens 2020;38:1878-89. [Crossref] [PubMed]
- Unudurthi SD, Luthra P, Bose RJC, et al. Cardiac inflammation in COVID-19: Lessons from heart failure. Life Sci 2020;260:118482. [Crossref] [PubMed]
- Liu Y, Liu Z, Tang H, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am J Physiol Cell Physiol 2019;317:C762-75. [Crossref] [PubMed]
- Mouton AJ, Li X, Hall ME, et al. Obesity, Hypertension, and Cardiac Dysfunction: Novel Roles of Immunometabolism in Macrophage Activation and Inflammation. Circ Res 2020;126:789-806. [Crossref] [PubMed]
- Huang JT, Liu SM. The role of N~6-methyladenosine modification in mRNA in clinical cancers. Chinese Bulletin of Life Sciences 2019;31:153-9. [Crossref]
- Kmietczyk V, Riechert E, Kalinski L, et al. m6A-mRNA methylation regulates cardiac gene expression and cellular growth. Life Sci Alliance 2019;2:e201800233. [Crossref] [PubMed]
- Murphy SP, Prescott MF, Camacho A, et al. Atrial Natriuretic Peptide and Treatment With Sacubitril/Valsartan in Heart Failure With Reduced Ejection Fraction. JACC Heart Fail 2021;9:127-36. [Crossref] [PubMed]
- Zhang B, Xu Y, Cui X, et al. Alteration of m6A RNA Methylation in Heart Failure With Preserved Ejection Fraction. Front Cardiovasc Med 2021;8:647806. [Crossref] [PubMed]
- Boissel S, Reish O, Proulx K, et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet 2009;85:106-11. [Crossref] [PubMed]
- Carnevali L, Graiani G, Rossi S, et al. Signs of cardiac autonomic imbalance and proarrhythmic remodeling in FTO deficient mice. PLoS One 2014;9:e95499. [Crossref] [PubMed]
- Gustavsson J, Mehlig K, Leander K, et al. FTO genotype, physical activity, and coronary heart disease risk in Swedish men and women. Circ Cardiovasc Genet 2014;7:171-7. [Crossref] [PubMed]
- Mathiyalagan P, Adamiak M, Mayourian J, et al. FTO-Dependent N6-Methyladenosine Regulates Cardiac Function During Remodeling and Repair. Circulation 2019;139:518-32. [Crossref] [PubMed]
- Zhihao L, Jingyu N, Lan L, et al. SERCA2a: a key protein in the Ca2+ cycle of the heart failure. Heart Fail Rev 2020;25:523-35. [Crossref] [PubMed]
- Hulot JS, Ishikawa K, Hajjar RJ. Gene therapy for the treatment of heart failure: promise postponed. Eur Heart J 2016;37:1651-8. [Crossref] [PubMed]
- Hubacek JA, Vymetalova J, Lanska V, et al. The fat mass and obesity related gene polymorphism influences the risk of rejection in heart transplant patients. Clin Transplant 2018;32:e13443. [Crossref] [PubMed]
- Zhang Y, Hamada M. DeepM6ASeq: prediction and characterization of m6A-containing sequences using deep learning. BMC Bioinformatics 2018;19:524. [Crossref] [PubMed]
- Sikorski V, Karjalainen P, Blokhina D, et al. Epitranscriptomics of Ischemic Heart Disease-The IHD-EPITRAN Study Design and Objectives. Int J Mol Sci 2021;22:6630. [Crossref] [PubMed]
- Mo X, Lei S, Zhang Y, et al. Genome-wide enrichment of m6A-associated single-nucleotide polymorphisms in the lipid loci. Pharmacogenomics J 2019;19:347-57. [Crossref] [PubMed]
- Cohain AT, Barrington WT, Jordan DM, et al. An integrative multiomic network model links lipid metabolism to glucose regulation in coronary artery disease. Nat Commun 2021;12:547. [Crossref] [PubMed]
- Mo XB, Lei SF, Zhang YH, et al. Examination of the associations between m6A-associated single-nucleotide polymorphisms and blood pressure. Hypertens Res 2019;42:1582-9. [Crossref] [PubMed]
- Slivnick J, Lampert BC. Hypertension and Heart Failure. Heart Fail Clin 2019;15:531-41. [Crossref] [PubMed]
- Kumari R, Dutta R, Ranjan P, et al. ALKBH5 Regulates SPHK1-Dependent Endothelial Cell Angiogenesis Following Ischemic Stress. Front Cardiovasc Med 2022;8:817304. [Crossref] [PubMed]
- Wu S, Zhang S, Wu X, et al. m6A RNA Methylation in Cardiovascular Diseases. Mol Ther 2020;28:2111-9. [Crossref] [PubMed]
- Wang T, Tian J, Jin Y. VCAM1 expression in the myocardium is associated with the risk of heart failure and immune cell infiltration in myocardium. Sci Rep 2021;11:19488. [Crossref] [PubMed]
- Mathoux J, Henshall DC, Brennan GP. Regulatory Mechanisms of the RNA Modification m6A and Significance in Brain Function in Health and Disease. Front Cell Neurosci 2021;15:671932. [Crossref] [PubMed]
- Jia G, Hill MA, Sowers JR. Diabetic Cardiomyopathy: An Update of Mechanisms Contributing to This Clinical Entity. Circ Res 2018;122:624-38. [Crossref] [PubMed]


