The Popeye domain containing genes: essential elements in heart rate control
The Popeye domain containing gene family
The search for novel genes with a cardiac-restricted gene expression pattern resulted in the identification of the first member of the Popeye domain containing genes Popdc1 (formerly named as Pop1) (1). The same gene was independently isolated by David Bader’s group and was named by them as Bves (2). Based on sequence homology and predicted protein structure, two additional family members were subsequently identified and named Popdc2 and Popdc3 (1).
All three Popdc genes display high level of expression in striated muscle tissue (skeletal muscle and heart). In addition, significant expression is found in smooth muscle tissue of bladder, uterus and the gastrointestinal tract (1,3-5). Popdc genes are also expressed in the central and autonomic nervous system and in epithelial cells of the epidermis, gastrointestinal tract, retina, lens, and cornea (3,6-9).
In the heart, atrial expression of Popdc1 is higher than in ventricle. Moreover the entire cardiac conduction system displays the most intense expression levels (5,10). The presence of Popdc1 in the epicardium and in epicardium-derived cells remains a debated and unresolved issue (1,4,5,11-14). Our own studies do not provide evidence for a significant expression in the epicardium or its derivatives. The related family member Popdc2 is expressed in the entire coelomic epithelium during early development, however, the epicardium and the coronary vasculature is devoid of expression in the embryonic and adult heart (3). Thus, we conclude that the major expression domain in the heart is in cardiac myocytes, which correlates well with the phenotypes observed in loss-of-function experiments (see below).
The evolution of the Popdc gene family
A hallmark of all Popdc genes is that they encode proteins with a large N-terminal hydrophobic domain, which is predicted to generate up to three membrane-spanning helices. Moreover, a conserved Popeye domain is present in all Popeye genes (Figure 1). The three Popdc genes are located on two different chromosomal loci. In man, POPDC1 and POPDC3 are organized in tandem on chromosome 6q21, with POPDC3 being upstream of POPDC1. Both genes are separated by about 20 kb of intergenic sequence. Interestingly, this locus appears to be transcribed from a bidirectional POPDC1 promoter and an antisense RNA starts in the intergenic region between POPDC1 and POPDC3 and extends to overlap the POPDC3 gene. It is currently unknown whether the antisense transcript affects POPDC3 expression, however, it is an attractive hypothesis given that POPDC3 is expressed at much lower level than POPDC1. Interestingly the intergenic region has grown significantly during evolution and is only 2 and 4 kb in tunicates and in zebrafish, respectively. In the chick, the intergenic distance has already risen to 8 kb. In this species and also predicted for the gene complex in bovines, a peculiar set of mRNAs have been observed containing sequences of both Popdc1 and Popdc3, suggesting that in these species, the boundaries between both genes are not strictly observed by the transcription apparatus (1). In tunicates, sea urchins, arthropodes (insects, spiders, and crustaceans) and molluscs, only two of the three Popdc genes are found, which appear to be orthologues of Popdc1 and Popdc3 and are clustered on the same chromosomal locus (Figure 1). Probably early in evolution a tandem-duplication occurred generating Popdc1 and Popdc3 and evolutionary pressure maintained them as a gene cluster probably in order to co-regulate and assure proper expression levels. It has been recently erroneously proposed that Popdc1 is the only gene present in invertebrates and vertebrates (16). This statement probably was based on the fact that in Drosophila only a single Popdc gene is present. However, the situation in Drosophila and other related species of the order Diptera (Anopheles) appears to be the exception rather than the rule. Interestingly in Daphnia pulex (water flea), a total of four different Popdc genes are found, one gene appears to be orthologous to Popdc1, whereas the other three are most similar to Popdc3. Popdc2 is uniquely present in vertebrates and appears to have arisen by gene duplication from Popdc3. Popdc2 and Popdc3 are more closely related to each other displaying an approximately 50% sequence identity on the protein level, while Popdc1 is only 25% similar to Popdc2 or Popdc3 (1). In man, Popdc2 is present on chromosome 3q13.33 and is the Popdc gene, which displays the highest expression level in the heart (Figure 1).
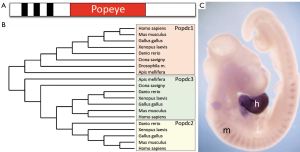 The Popeye gene family. A. Schematic depiction of the structure of the Popdc proteins. Members of the Popdc gene family encode membrane proteins, which have up to three transmembrane domains (black bars). In addition an evolutionary conserved Popeye domain is present; B. Dendogram of the Popeye gene family in vertebrate and invertebrate species. In vertebrates three genes are identified, Popdc1, Popdc2, and Popdc3. In lower chordates (Ciona savigny) and in insects (Apis mellifera) two genes are found, which are probably orthologus to Popdc1 and Popdc3. In Drosophila and probably all Diptera only a single Popdc gene is found. Popdc2 is only present in vertebrates. The dendrogram was generated with the help of the Phylogeny.fr program (15); C. Popdc genes are preferentially expressed in muscle and heart tissue. Depicted here is a three days old chick embryo, which was subjected to whole mount in situ hybridization detecting Popdc2 in the heart (h) and myotome (m), which are embryonic structures that give rise to the back muscles
The Popeye gene family. A. Schematic depiction of the structure of the Popdc proteins. Members of the Popdc gene family encode membrane proteins, which have up to three transmembrane domains (black bars). In addition an evolutionary conserved Popeye domain is present; B. Dendogram of the Popeye gene family in vertebrate and invertebrate species. In vertebrates three genes are identified, Popdc1, Popdc2, and Popdc3. In lower chordates (Ciona savigny) and in insects (Apis mellifera) two genes are found, which are probably orthologus to Popdc1 and Popdc3. In Drosophila and probably all Diptera only a single Popdc gene is found. Popdc2 is only present in vertebrates. The dendrogram was generated with the help of the Phylogeny.fr program (15); C. Popdc genes are preferentially expressed in muscle and heart tissue. Depicted here is a three days old chick embryo, which was subjected to whole mount in situ hybridization detecting Popdc2 in the heart (h) and myotome (m), which are embryonic structures that give rise to the back muscles
There is ample evidence that Popdc1/Popdc3 are co-regulated and differ in their expression level and pattern from Popdc2. For example it has been reported that POPDC1 and POPDC3 are down-regulated in the failing human heart (17). Correlation analyses of mRNA levels among the failing heart specimens revealed a coordinated regulation of POPDC1 with POPDC3, which differed from POPDC2. Similarly it was found that POPDC1 and POPDC3 but not POPDC2 are hypermethylated in gastric and colorectal tumor specimen (6,7).
The popeye domain binds cAMP
Popdc genes encode membrane proteins, which are abundantly present in the plasma membrane of cardiac myocytes (Figure 2). The protein is evenly distributed in both intercalated disk and lateral membranes and is also present in the T-tubules of ventricular myocytes. The three vertebrate Popdc genes encode proteins, which are extensively glycosylated, localized to the plasma membrane, and are between 300 and 360 amino acids long. The membrane topology has been extensively studied in case of Popdc1 and the amino terminus is extracellular, while the carboxy terminus is cytoplasmic (18). Popdc proteins have a short extracellular amino terminus of approx. 25-40 amino acids, harboring one or two N-glycosylation sites followed by three transmembrane domains (Figure 2). In the cytoplasm a highly conserved Popeye domain is found which functions as a cyclic nucleotide-binding domain and is followed by a terminal sequence, which is variable in length and subject to alternative splicing. Popdc1 has been shown to homodimerize (18) and a sequence motif, which is essential for dimerization and unique to Popdc1 has been mapped to the carboxy-terminal part of the Popeye domain (19), however recent data suggest that other domains in the molecule also contribute to dimerization (20).
The Popeye domain shows the highest level of sequence conservation within the Popdc proteins, suggesting that this protein domain is probably of biological importance. Structural modeling revealed that it is similar to the cAMP-binding domain of protein kinase A (PKA) (Figure 2) (10). However, the amino acid sequence of the putative cyclic nucleotide-binding cassette (CNBC) did not match the sequences found in other CNBCs (Figure 2). The ability to bind cAMP was tested using affinity precipitation with cAMP agarose and by radio-ligand binding assay. Both assays demonstrated high-affinity binding of cAMP in the physiological relevant range (10). The ability to bind cAMP is shared by Popdc2 and Popdc3, which is not surprising given the high level of structural and sequence conservation of the Popeye domain. There was roughly a 20-fold difference in the affinity for cAMP and cGMP using a recombinant Popeye domain of Popdc1, suggesting preferential binding of Popdc1 to cAMP. It has been reported for PKA and PKG that these kinases can bind both nucleotides although with quantitative differences. It only requires the change of one amino acid to convert PKA into an enzyme which gets activated equally well by both nucleotides (21). Thus it is possible that at very high concentrations of cGMP this nucleotide is also able to bind to Popdc proteins. In FRET experiments, however a complete insensitivity of Popdc1 to nitroprusside-induced changes in cGMP levels was observed, suggesting that under physiological conditions Popdc proteins will bind and mediate only cAMP-induced signal transduction (10).
Overall, the Popeye domain reveals high sequence conservation. There are several amino acids that are invariant and present in all Popeye domains from oyster to man. The projection of the conserved amino acids onto the predicted 3-dimensional structure revealed a conserved cluster around and including the CNBC. In order to test whether evolutionary conserved residues are functionally important, the highly conserved peptide motifs DSPE and FQVT (Figure 2) were mutagenized. In particular charge-to-alanine mutations at position D200 in Popdc1 and D184 in Popdc2 resulted in a dramatic reduction in cAMP-binding suggesting that an aspartate residue at this position is essential for cyclic nucleotide-binding (10). Unfortunately, until now no structural information of the Popeye domain is available, which however will be essential to further define the interaction of the Popeye domain with cyclic nucleotides.
Interestingly, the protein with the highest level of sequence conservation of a non-Popdc protein is the cyclic AMP receptor protein (CRP), a bacterial transcription factor, which regulates enzyme expression in response to alterations in metabolite concentration. Thus, it is possible that the Popdc proteins have actually evolved from a transcriptional regulator and have gained a novel subcellular localization at the plasma membrane. In this regard it is noteworthy that Popdc proteins are also found in an isoform-dependent manner in the nucleus of both cardiac and skeletal muscle cells, which would suggest that the proteins may have retained some of their ancestral function (22).
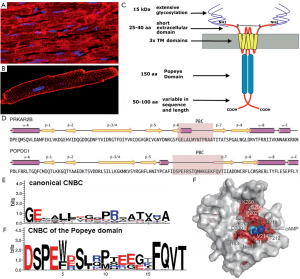 Figure 2. Popdc proteins are membrane proteins that bind cAMP. (A,B) Immunostaining of Popdc1 in (A) adult mouse myocardium and (B) isolated adult mouse ventricular myocyte. Popdc1 is localized at the intercalated disk region, lateral membranes, and in the T-tubular system. (C) Working model of Popdc1 protein. The protein consists of a short extracellular domain, three transmembrane domains and an evolutionary conserved Popeye domain followed by a carboxy-terminal region, which is variable in length and sequence. Note that Popdc1 is found as a dimer at the plasma membrane and a disulfide bridge stabilizes protein dimerization. (D) Secondary structure prediction of human POPDC1, displaying structural similarity to the cAMP-binding domain of protein kinase A (PRKAR2B). (E,F) Comparison of the sequence of the cyclic nucleotide binding cassette (CNBC) found in (E) established vertebrate cAMP-binding proteins, and (F) the presumptive CNBC in Popeye proteins. Despite both sequences bind cAMP with high affinity, the CNBCs do not resemble each other. (G) 3-D model of the Popeye domain of POPDC1. Amino acid residues labeled in red are invariable and mostly cluster around the presumptive CNBC. Panel D and G are reproduced from (10) with permission
Figure 2. Popdc proteins are membrane proteins that bind cAMP. (A,B) Immunostaining of Popdc1 in (A) adult mouse myocardium and (B) isolated adult mouse ventricular myocyte. Popdc1 is localized at the intercalated disk region, lateral membranes, and in the T-tubular system. (C) Working model of Popdc1 protein. The protein consists of a short extracellular domain, three transmembrane domains and an evolutionary conserved Popeye domain followed by a carboxy-terminal region, which is variable in length and sequence. Note that Popdc1 is found as a dimer at the plasma membrane and a disulfide bridge stabilizes protein dimerization. (D) Secondary structure prediction of human POPDC1, displaying structural similarity to the cAMP-binding domain of protein kinase A (PRKAR2B). (E,F) Comparison of the sequence of the cyclic nucleotide binding cassette (CNBC) found in (E) established vertebrate cAMP-binding proteins, and (F) the presumptive CNBC in Popeye proteins. Despite both sequences bind cAMP with high affinity, the CNBCs do not resemble each other. (G) 3-D model of the Popeye domain of POPDC1. Amino acid residues labeled in red are invariable and mostly cluster around the presumptive CNBC. Panel D and G are reproduced from (10) with permission
The Popdc proteins interact with the two-pore channel TREK-1
The localization of Popdc proteins at the plasma membrane is suggestive of an interaction with membrane proteins. Since cardiac arrhythmias are present in null mutant mice and zebrafish morphants (see below), we hypothesized that Popdc proteins might interact with cardiac ion channels or pumps. A screen was set up in the Xenopus oocyte system in order to reveal current changes in the presence of Popdc1 protein. Interestingly, a specific interaction of the potassium channel TREK-1 with Popdc1 was observed (10). TREK-1 current was 2-fold increased in the presence of Popdc1 and this was also seen for the other members of the Popdc family. This increase was based on an enhanced membrane representation of TREK-1, suggesting a modulation of channel trafficking by Popdc proteins.
TREK-1 is a potassium background channel, which is important for setting the resting membrane potential in neuronal cells (23) and belongs to the family of two-pore potassium (K2P) channels of which at least three other channels, TASK-1, TREK-2 and TWIK-2 are expressed in the heart (24), however, only TREK-1 displayed a specific interaction with Popdc1 (10). The specific interaction of Popdc1 with TREK-1 was utilized to generate a bi-molecular FRET assay, which corroborated the specific interaction between both proteins. Protein interaction was sensitive to adrenergic stimulation. A rapid reduction of the FRET signal following isoproterenol stimulation shows that the Popdc1-TREK-1 interaction is sensitive to changes in cAMP levels. Modulation of the FRET signal was only induced by raising cAMP levels and was not observed in response to cGMP (10). Since TREK-1 is subject to PKA-dependent phosphorylation and inactivation, the Popdc1-dependent modulation of TREK-1 current may represent a fast response to adrenergic stimulation followed by the slower kinase-dependent inactivation (25). Due to the lack of specific antagonist it has been difficult to assign specific roles to this channel in the cardiovascular system. It has been proposed that TREK-1 may be involved in mechano-electrical feedback regulation (26,27). TREK-1 may also be involved in modulating natriuretic peptide secretion by atrial myocardium (28). Last but not least the related K2P ion channel ORK1 in Drosophila has been implicated in regulating pacemaker activity (29). Since the role of TREK-1 in the cardiovascular system has not been fully elucidated, the functional significance of the interaction with Popdc proteins has not been fully established. It is however likely, given the abundance of Popdc proteins in cardiac myocytes, that in addition to TREK-1 other membrane proteins will also be interaction partners of Popdc proteins.
Functional analysis of Popdc1 and Popdc2 in zebrafish and mice
As in other vertebrates, zebrafish popdc2 is expressed in skeletal muscle and heart (30) (Figure 3). Loss-of-function analysis of popdc2 by morpholino treatment of zebrafish embryos revealed a severe muscle defect, which is characterized by myofiber alignment defects and rupture due to an aberrant structure of the myotendinous junction (30). In addition, the craniofacial muscles were severely reduced in size. The observation of a defective skeletal muscle development is reminiscent of the muscle regeneration phenotype observed in the Popdc1 null mutant in mice (14). The heart in popdc2 morphants displayed massive pericardial edema, which is an indication for embryonic heart failure (30). Reduction of the morpholino concentration normalized the morphology of the developing skeletal muscle and heart. However, a bradyarrhythmia developed between day 4 and day 7 post fertilization. The heart rate was significantly reduced and hearts with a partial or complete AV block were present in many morphants (Figure 3). In some embryos also a sinuatrial block was observed.
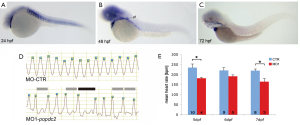 Figure 3. Loss of popdc2 in zebrafish causes cardiac arrhythmia. (A-C) Expression of popdc2 in (A) 1-day, (B) 2-day, and (C) 3-day old zebrafish embryos, which are expressed in the heart and skeletal muscle. (D) Ventricular contractions injected with control (M-CTR) and popdc2 (MO1-popdc2) morpholinos. (E) Mean heart rates of control and popdc2 morpholino-injected embryos reveal a bradycardia in the morphants. Panel A-E are reproduced from (30) with permission
Figure 3. Loss of popdc2 in zebrafish causes cardiac arrhythmia. (A-C) Expression of popdc2 in (A) 1-day, (B) 2-day, and (C) 3-day old zebrafish embryos, which are expressed in the heart and skeletal muscle. (D) Ventricular contractions injected with control (M-CTR) and popdc2 (MO1-popdc2) morpholinos. (E) Mean heart rates of control and popdc2 morpholino-injected embryos reveal a bradycardia in the morphants. Panel A-E are reproduced from (30) with permission In order to gain insight into the role of the Popdc genes in mammals, null mutations were engineered in mice by knockin of a beta-galactosidase reporter into the first coding exon of Popdc1 and Popdc2 (10). Both mutants were born at Mendelian frequency suggesting no embryonic lethality. The LacZ reporter genes were used to study the expression of Popdc1 and -2 in further detail. A strong expression of both genes is observed in cardiac myocytes, while non-muscle cells in the heart do not express Popdc1 or Popdc2 (Figure 4) (10). For both genes a high level of expression was observed in the cardiac conduction system (CCS). The sinoatrial node (SAN), the atrioventricular node (AVN), the His bundle, and the Purkinje fibres were strongly labeled by LacZ staining and express high levels of both, Popdc1 and Popdc2 (Figure 4). There are also differences in the expression pattern between Popdc1 and Popdc2. In particular the expression of Popdc1 in the ventricular working myocardium is significantly lower than in the atria, while Popdc2 appears to be homogenously expressed in both chamber types (Figure 4).
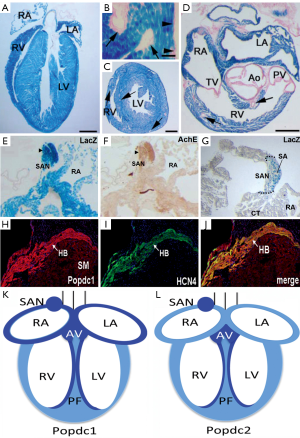 Figure 4. Popdc1 and Popdc2 are expressed in working myocardium and the cardiac conduction system (CCS). (A-D) LacZ staining of sections through the heart of an adult Popdc2LacZ mouse. (A) Frontal section of an adult heart demonstrating expression in the different cardiac chambers. (B-D) Transverse sections through the ventricular region demonstrating strong expression in the cardiac myocytes and the absence in the (B,C) coronary vasculature (arrows), epicardium (arrowheads), and (D) valve tissue. (E-G) Strong expression of Popdc2LacZ in the sinoatrial node (SAN). (E,F) Consecutive sections were stained for (E) LacZ and (F) acetylcholine esterase. (G) Transverse section through the vena cava region demonstrating high level of expression in SAN myocytes. (H-J) Immunohistochemical detection of (H) Popdc1, (I) HCN4, and (J) merge of the two stainings in the His bundle (HB) of an adult mouse heart. Note the relative high level of expression in the HB in comparison to the significantly lower level in working myocardium (WM). (K,L) Graphic summary of the expression domains of Popdc1 and Popdc2 in the adult mouse heart. Both genes are strongly expressed in the cardiac conduction system. (K) Popdc1 shows a higher expression level in the atria than in the ventricles. (L) Popdc2 display robust expression in the atrial and ventricular chambers. Panel (A-D) are reproduced from (3) with permission
Figure 4. Popdc1 and Popdc2 are expressed in working myocardium and the cardiac conduction system (CCS). (A-D) LacZ staining of sections through the heart of an adult Popdc2LacZ mouse. (A) Frontal section of an adult heart demonstrating expression in the different cardiac chambers. (B-D) Transverse sections through the ventricular region demonstrating strong expression in the cardiac myocytes and the absence in the (B,C) coronary vasculature (arrows), epicardium (arrowheads), and (D) valve tissue. (E-G) Strong expression of Popdc2LacZ in the sinoatrial node (SAN). (E,F) Consecutive sections were stained for (E) LacZ and (F) acetylcholine esterase. (G) Transverse section through the vena cava region demonstrating high level of expression in SAN myocytes. (H-J) Immunohistochemical detection of (H) Popdc1, (I) HCN4, and (J) merge of the two stainings in the His bundle (HB) of an adult mouse heart. Note the relative high level of expression in the HB in comparison to the significantly lower level in working myocardium (WM). (K,L) Graphic summary of the expression domains of Popdc1 and Popdc2 in the adult mouse heart. Both genes are strongly expressed in the cardiac conduction system. (K) Popdc1 shows a higher expression level in the atria than in the ventricles. (L) Popdc2 display robust expression in the atrial and ventricular chambers. Panel (A-D) are reproduced from (3) with permission Stress-induced bradycardia phenotype in Popdc1 and Popdc2
Due to the high expression level in the CCS for both genes, the phenotypic consequences of a knockout of Popdc1 and Popdc2 for these tissues was assessed by Electrocardiogram (ECG) analysis in conscious animals (10). After subjecting mutants to physical or mental stress, they displayed high variability in their heart frequencies, which was due to sinus pauses (Figure 5). Under baseline conditions no differences in heart rate and no sinus pauses were recorded.
Such a stress-induced bradycardia phenotype is reminiscent of the sick sinus syndrome (SSS), or sinus node disease (SND) which can manifest as sinus bradycardia, sinus arrest and/or sino-atrial block and is the most frequent indication for pacemaker implantation (31). SSS is most prevalent in the elderly in the absence of apparent accompanying heart disease (32). Intriguingly, the sinus bradycardia in Popdc mutants was age-dependent as well. In young adults (3 months of age) no sinus bradycardia were observed, at 5.5 months of age there was an increase in sinus pauses in response to stress but the phenotype wasn’t fully penetrant, while at 8 month all mutants had severe sinus bradycardia due to a high number of sinus pauses (Figure 5).
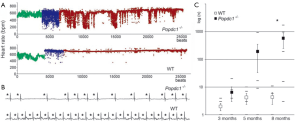 Figure 5. Popdc null mutants have an age-dependent stress-induced sinus bradycardia. Popdc1 mutants were implanted with telemetric ECG devices. Subsequently they were subjected to a 5 min swimming exercise. A. Heart rate (y axis) of Popdc1–/– and wild type (WT) mice before (green), during (blue), and after (red) swim stress, depicted on a beat per beat basis (x axis); B. ECG recordings of Popdc1–/– and WT mice during swim stress test. Asterisks indicate P waves. Horizontal bar: 100 ms. Note the high number of sinus pauses in the mutant animal; C. Number of pauses in WT and Popdc1–/– mice during a 30-minute period after swim stress as a function of age. *P<0.05 between genotypes. Note that at the age of 3 and 6 months no significant differences in the number of pauses were observed. However at 8 months of age a fully penetrant phenotype was present. An identical phenotype was observed for the Popdc2 null mutant. Panel A-C are reproduced from (10) with permission
Figure 5. Popdc null mutants have an age-dependent stress-induced sinus bradycardia. Popdc1 mutants were implanted with telemetric ECG devices. Subsequently they were subjected to a 5 min swimming exercise. A. Heart rate (y axis) of Popdc1–/– and wild type (WT) mice before (green), during (blue), and after (red) swim stress, depicted on a beat per beat basis (x axis); B. ECG recordings of Popdc1–/– and WT mice during swim stress test. Asterisks indicate P waves. Horizontal bar: 100 ms. Note the high number of sinus pauses in the mutant animal; C. Number of pauses in WT and Popdc1–/– mice during a 30-minute period after swim stress as a function of age. *P<0.05 between genotypes. Note that at the age of 3 and 6 months no significant differences in the number of pauses were observed. However at 8 months of age a fully penetrant phenotype was present. An identical phenotype was observed for the Popdc2 null mutant. Panel A-C are reproduced from (10) with permission Recent investigations have linked genetic ion channel defects with familial SSS including mutations in the sodium channel SCN5A (33) and HCN4, the gene encoding the hyperpolarization-activated cyclic nucleotide gated channel generating the “funny” pacemaker current in SAN cells. The observation that in both Popdc null mutants a SSS-like phenotype exists suggests that these genes may be novel candidate genes for this disease and mutations might be found in familial cases. In addition, both HCN4 and SCN5A are candidate interaction partners for Popdc proteins. However If is not affected in mutant SAN both at baseline and after 8-Br-cAMP treatment (10).
It is likely that by defining the role of Popdc proteins in SAN pacemaker function novel insight will be gained into pacemaker function and possibly also the underlying causes for SSS. Pacemaking involves spontaneous diastolic depolarization, which has been linked to voltage-sensitive membrane currents, such as the hyperpolarization-activated pacemaker current If (Figure 6). If depolarizes the membrane to a level sufficient for the L-type Ca2+-channel to open and to initiate an action potential. Support for a major role of If has been obtained by the fact that mutations in HCN4 causes a reduced basal heart rate (34) and likewise drugs that block If such as Ivabradine have a similar effect (35). Surprisingly, however, point mutations of HCN4 are associated with baseline bradycardia but have no effect on exercise (36). Similarly, a mutation in the cAMP-binding domain of HCN4 causes an embryonic lethal phenotype in transgenic mice but no effect is seen on stress-inducibility of cardiac pacemaking in the adult (37). Another mouse mutant with heart-specific and inducible expression of a human HCN4 mutation abolishing the cAMP-dependent regulation of HCN channels did not affect heart rate adaptation during physical activity (38). A similar conclusion was also drawn by yet another mouse mutant with a SAN-specific deletion of HCN4 (39). It has been proposed that in the adult heart HCN4 acts as a depolarization reserve but it is not essential for heart rate adaptation in response to stress.
Alternatively to voltage-sensitive ion channels, which collectively represent the voltage or membrane clock, there is evidence for another oscillatory mechanism in the pacemaker cells. The spontaneous local subsarcolemmal Ca2+ release (LCR) from the sarcoplasmatic reticulum was observed in sinoatrial myocytes and was termed the Ca2+ clock (Figure 6) (40). Evidence for an important role of the Ca2+ clock stems from the observation that ryanodine treatment causes a slowing of the cardiac pacemaker (41). The LCR occurs in the sinuatrial node cell independently of the membrane voltage and drives a net inward sodium current due to the activity of the Na+-Ca2+-exchanger (NCX), which therefore represents a link between both clocks. Adrenergic stimulation causes increased protein kinase A activity, which results in phosphorylation and activity modulation of a number of proteins being part of the membrane and voltage clock networks. In addition, CaMKII has also been implicated as another mediator of pacemaker adaptation to stress (42). A key question for the future will be to understand the role of Popdc proteins stress-induced pacemaker adaptation. In particular its role in the context of the membrane and calcium clocks needs to be analyzed, which may help to explain the stress-induced pauses in the Popdc1 and Popdc2 null mutants. It is possible that the ability of Popdc proteins to bind cAMP is an important input to modulate the activity of the complex pacemaker network.
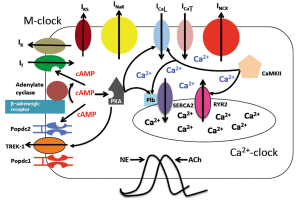 Figure 6. Popdc proteins are probably part of the regulatory network controlling cardiac pacemaking. A complex network of membrane localized voltage-gated ion channels, collectively named the membrane clock (M-clock) and the Ca2+-clock, which consists of the oscillatory local Ca2+-release from the sarcoplasmatic reticulum via the ryanodine receptor (RYR2) and re-uptake by SERCA2, are interlinked by the Na-Ca2+-exchanger (NCX), which causes an inward sodium current in response to a local Ca2+-release. Several inward currents, If and the L- and T-type Ca2+-channels drive the depolarisation of the pacemaker cell, while potassium currents are involved in repolarization. A unique property of the pacemaker cell is the lack of a resting potential due to the opening of the hyperpolarization-activated cyclic nucleotide-gated (HCN) channels at a hyperpolarizing membrane potential. Adrenergic stimulation results in an increased heart frequency due to a faster pacemaker rate. The binding of cAMP causes an activation of protein kinase A, which phosphorylates phospholamban and the Ca2+-channels, resulting in an increase in the Ca2+-inward current and Ca2+- uptake. In addition the HCN channels have also a cAMP-binding domain. The role of the Popdc proteins in this context is currently not well understood. While the two-pore channel TREK-1 is an established interaction partner for both Popdc1 and Popdc2, its role in cardiac pacemaking has not been elucidated. It is likely that other proteins (possibly channels or pumps participating in the membrane and Ca2+-clock) are also interacting with Popdc proteins. The significant finding of the presence of SAN pauses in response to physiological stress suggest that Popdc proteins are involved in the adaptation of pacemaking in response to increased levels of cAMP. Another element in the complex network is CaMKII, which phosphorylates several critical proteins of the pacemaking network
Figure 6. Popdc proteins are probably part of the regulatory network controlling cardiac pacemaking. A complex network of membrane localized voltage-gated ion channels, collectively named the membrane clock (M-clock) and the Ca2+-clock, which consists of the oscillatory local Ca2+-release from the sarcoplasmatic reticulum via the ryanodine receptor (RYR2) and re-uptake by SERCA2, are interlinked by the Na-Ca2+-exchanger (NCX), which causes an inward sodium current in response to a local Ca2+-release. Several inward currents, If and the L- and T-type Ca2+-channels drive the depolarisation of the pacemaker cell, while potassium currents are involved in repolarization. A unique property of the pacemaker cell is the lack of a resting potential due to the opening of the hyperpolarization-activated cyclic nucleotide-gated (HCN) channels at a hyperpolarizing membrane potential. Adrenergic stimulation results in an increased heart frequency due to a faster pacemaker rate. The binding of cAMP causes an activation of protein kinase A, which phosphorylates phospholamban and the Ca2+-channels, resulting in an increase in the Ca2+-inward current and Ca2+- uptake. In addition the HCN channels have also a cAMP-binding domain. The role of the Popdc proteins in this context is currently not well understood. While the two-pore channel TREK-1 is an established interaction partner for both Popdc1 and Popdc2, its role in cardiac pacemaking has not been elucidated. It is likely that other proteins (possibly channels or pumps participating in the membrane and Ca2+-clock) are also interacting with Popdc proteins. The significant finding of the presence of SAN pauses in response to physiological stress suggest that Popdc proteins are involved in the adaptation of pacemaking in response to increased levels of cAMP. Another element in the complex network is CaMKII, which phosphorylates several critical proteins of the pacemaking network The only established ion channel with which Popdc proteins are interacting is TREK-1 (10). TREK-1 current generates an outward potassium current causing hyperpolarization of the cell membrane rendering the cell less excitable. While its role in setting the resting membrane potential is fully established in neurons, its role in cardiac pacemaking in vertebrates has not been investigated. TREK-1 channels may reduce the ability of sinus node cells to reach the threshold for activation. The absence of Popdc1 or Popdc2 should lead to a reduction of TREK-1 current and as result causing an increase in sinus node activity, however, the reverse is observed in the both mouse mutants. Therefore the sinus pauses that were observed in the Popdc mutants cannot be explained on the basis of an inhibition of TREK-1 current alone but may involve other interaction partners yet to be discovered.
Concluding remarks
This review focused on the role of the Popdc gene family in the heart. Clearly there are additional expression domains, suggesting that other organs also require Popdc gene function (16). Indeed, loss of function analysis of Popdc1 in Drosophila (43), Xenopus (44), and zebrafish (45) indicated an early requirement for this gene during gastrulation. All three knockdown studies described impaired cell migration and epithelium formation. However antisense oligonucleotides and morpholinos were employed, which could be subject to off-target effects. In this regard it needs to be stressed that an early embryonic lethality has not been observed in the Popdc1 mouse mutant (46). However, our analysis so far indicates that there is extensive overlap in gene function and expression, which may explain why the mouse embryo is protected from an early embryonic lethality. Drosophila has only a single Popdc gene and this may explain why an early embryonic phenotype is observed.
Functions of Popdc genes in epithelium formation and function is also suggested by the finding that a loss-of Popdc1 function impairs tight junction formation (47) and signaling (20). It has also been suggested that cell migration and cell adhesion are affected by a loss of Popdc1, which is probably due to the interaction of Popdc1 with GEFT that modulates Rac1/Cdc42 activity (48) and Vamp3, which is involved in β-integrin mediated cell adhesion (49). Consistent with these studies are the recent reports that silencing of POPDC1 and POPDC3 by promoter hypermethylation is associated with colorectal carcinoma (50) and gastric cancer (6). Moreover, abnormal expression of POPDC1 and POPDC3 has a strong impact on the survival rate of gastric cancer patients (51). Another cancer in which POPDC1 was found to be hypermethylated is non small cell lung cancer (52). Similar to the case of gastric cancer, it is believed that POPDC1 levels in the transformed cells determine the progression of carcinogenesis but hypermethylation is not observed in the precancerous state (53).
A study on the expression levels of Popdc genes in heart failure reported a specific loss of POPDC1 and POPDC3 in a large fraction of patients (17). Given the important role regulating excitation in cardiac myocytes it is possible that the loss of this genes in the failing heart will increase the risk of developing cardiac arrhythmia and sudden cardiac death, two conditions which are prevalent in heart failure patients (54).
Our understanding of Popdc gene function and its association with cardiac arrhythmias and tumor progression has just started to emerge. It will be important to identify additional Popdc-interacting proteins, which may help to unravel the molecular functions of this exciting gene family and its association with heart disease.
Acknowledgements
The authors gratefully acknowledge the financial support by the German Research council, the Medical Research Council (MRC), Imperial College London, and the Magdi Yacoub Research Foundation, who funded the research in the author’s laboratory.
Disclosure: The authors declare that this manuscript has not been submitted elsewhere and disclose absence of any financial interest or association with a company.
References
- Andrée B, Hillemann T, Kessler-Icekson G, et al. Isolation and characterization of the novel popeye gene family expressed in skeletal muscle and heart. Dev Biol 2000;223:371-82.
- Reese DE, Zavaljevski M, Streiff NL, et al. bves: A novel gene expressed during coronary blood vessel development. Dev Biol 1999;209:159-71.
- Froese A, Brand T. Expression pattern of Popdc2 during mouse embryogenesis and in the adult. Dev Dyn 2008;237:780-7.
- Breher SS, Mavridou E, Brenneis C, et al. Popeye domain containing gene 2 (Popdc2) is a myocyte-specific differentiation marker during chick heart development. Dev Dyn 2004;229:695-702.
- Torlopp A, Breher SS, Schlüter J, et al. Comparative analysis of mRNA and protein expression of Popdc1 (Bves) during early development in the chick embryo. Dev Dyn 2006;235:691-700.
- Kim M, Jang HR, Haam K, et al. Frequent silencing of popeye domain-containing genes, BVES and POPDC3, is associated with promoter hypermethylation in gastric cancer. Carcinogenesis 2010;31:1685-93.
- Williams CS, Zhang B, Smith JJ, et al. BVES regulates EMT in human corneal and colon cancer cells and is silenced via promoter methylation in human colorectal carcinoma. J Clin Invest 2011;121:4056-69.
- Ripley AN, Chang MS, Bader DM. Bves is expressed in the epithelial components of the retina, lens, and cornea. Invest Ophthalmol Vis Sci 2004;45:2475-83.
- Smith TK, Bader DM. Characterization of Bves expression during mouse development using newly generated immunoreagents. Dev Dyn 2006;235:1701-8.
- Froese A, Breher SS, Waldeyer C, et al. Popeye domain containing proteins are essential for stress-mediated modulation of cardiac pacemaking in mice. J Clin Invest 2012;122:1119-30.
- Wada AM, Reese DE, Bader DM. Bves: prototype of a new class of cell adhesion molecules expressed during coronary artery development. Development 2001;128:2085-93.
- Wada AM, Smith TK, Osler ME, et al. Epicardial/Mesothelial cell line retains vasculogenic potential of embryonic epicardium. Circ Res 2003;92:525-31.
- Osler ME, Smith TK, Bader DM. Bves, a member of the Popeye domain-containing gene family. Dev Dyn 2006;235:586-93.
- Andrée B, Fleige A, Arnold HH, et al. Mouse Pop1 is required for muscle regeneration in adult skeletal muscle. Mol Cell Biol 2002;22:1504-12.
- Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 2008;36:W465-9.
- Hager HA, Bader DM. Bves: ten years after. Histol Histopathol 2009;24:777-87.
- Gingold-Belfer R, Bergman M, Alcalay Y, et al. Popeye domain-containing 1 is down-regulated in failing human hearts. Int J Mol Med 2011;27:25-31.
- Knight RF, Bader DM, Backstrom JR. Membrane topology of Bves/Pop1A, a cell adhesion molecule that displays dynamic changes in cellular distribution during development. J Biol Chem 2003;278:32872-9.
- Kawaguchi M, Hager HA, Wada A, et al. Identification of a novel intracellular interaction domain essential for Bves function. PLoS One 2008;3:e2261.
- Russ PK, Pino CJ, Williams CS, et al. Bves modulates tight junction associated signaling. PLoS One 2011;6:e14563.
- Shabb JB, Ng L, Corbin JD. One amino acid change produces a high affinity cGMP-binding site in cAMP-dependent protein kinase. J Biol Chem 1990;265:16031-4.
- Schindler R, Simrick S, Brand T. Nuclear localization of members of the popeye domain containing (Popdc) protein family. Cardiovasc Res 2012;93:S98.
- Noël J, Sandoz G, Lesage F. Molecular regulations governing TREK and TRAAK channel functions. Channels (Austin) 2011;5:402-9.
- Liu W, Saint DA. Heterogeneous expression of tandem-pore K+ channel genes in adult and embryonic rat heart quantified by real-time polymerase chain reaction. Clin Exp Pharmacol Physiol 2004;31:174-8.
- Terrenoire C, Lauritzen I, Lesage F, et al. A TREK-1-like potassium channel in atrial cells inhibited by beta-adrenergic stimulation and activated by volatile anesthetics. Circ Res 2001;89:336-42.
- Kelly D, Mackenzie L, Hunter P, et al. Gene expression of stretch-activated channels and mechanoelectric feedback in the heart. Clin Exp Pharmacol Physiol 2006;33:642-8.
- Xian Tao Li, Dyachenko V, Zuzarte M, et al. The stretch-activated potassium channel TREK-1 in rat cardiac ventricular muscle. Cardiovasc Res 2006;69:86-97.
- Ogawa T, Forero M, Burgon PG, et al. Role of potassium channels in stretch-promoted atrial natriuretic factor secretion. J Am Soc Hypertens 2009;3:9-18.
- Lalevée N, Monier B, Sénatore S, et al. Control of cardiac rhythm by ORK1, a Drosophila two-pore domain potassium channel. Curr Biol 2006;16:1502-8.
- Kirchmaier BC, Poon KL, Schwerte T, et al. The Popeye domain containing 2 (popdc2) gene in zebrafish is required for heart and skeletal muscle development. Dev Biol 2012;363:438-50.
- Bernstein AD, Parsonnet V. Survey of cardiac pacing in the United States in 1989. Am J Cardiol 1992;69:331-8.
- Dobrzynski H, Boyett MR, Anderson RH. New insights into pacemaker activity: promoting understanding of sick sinus syndrome. Circulation 2007;115:1921-32.
- Benson DW, Wang DW, Dyment M, et al. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A). J Clin Invest 2003;112:1019-28.
- Schulze-Bahr E, Neu A, Friederich P, et al. Pacemaker channel dysfunction in a patient with sinus node disease. J Clin Invest 2003;111:1537-45.
- Bucchi A, Barbuti A, Baruscotti M, et al. Heart rate reduction via selective ‘funny’ channel blockers. Curr Opin Pharmacol 2007;7:208-13.
- Nof E, Luria D, Brass D, et al. Point mutation in the HCN4 cardiac ion channel pore affecting synthesis, trafficking, and functional expression is associated with familial asymptomatic sinus bradycardia. Circulation 2007;116:463-70.
- Harzheim D, Pfeiffer KH, Fabritz L, et al. Cardiac pacemaker function of HCN4 channels in mice is confined to embryonic development and requires cyclic AMP. EMBO J 2008;27:692-703.
- Alig J, Marger L, Mesirca P, et al. Control of heart rate by cAMP sensitivity of HCN channels. Proc Natl Acad Sci U S A 2009;106:12189-94.
- Herrmann S, Stieber J, Stöckl G, et al. HCN4 provides a ‘depolarization reserve’ and is not required for heart rate acceleration in mice. EMBO J 2007;26:4423-32.
- Lakatta EG, Maltsev VA, Vinogradova TM. A coupled SYSTEM of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart’s pacemaker. Circ Res 2010;106:659-73.
- Bogdanov KY, Maltsev VA, Vinogradova TM, et al. Membrane potential fluctuations resulting from submembrane Ca2+ releases in rabbit sinoatrial nodal cells impart an exponential phase to the late diastolic depolarization that controls their chronotropic state. Circ Res 2006;99:979-87.
- Wu Y, Gao Z, Chen B, et al. Calmodulin kinase II is required for fight or flight sinoatrial node physiology. Proc Natl Acad Sci U S A 2009;106:5972-7.
- Lin S, Zhao D, Bownes M. Blood vessel/epicardial substance (bves) expression, essential for embryonic development, is down regulated by Grk/EFGR signalling. Int J Dev Biol 2007;51:37-44.
- Ripley AN, Osler ME, Wright CV, et al. Xbves is a regulator of epithelial movement during early Xenopus laevis development. Proc Natl Acad Sci U S A 2006;103:614-9.
- Wu YC, Liu CY, Chen YH, et al. Blood Vessel Epicardial Substance (Bves) Regulates Epidermal Tight Junction Integrity through Atypical Protein Kinase C. J Biol Chem 2012;287:39887-97.
- Andrée B, Hillemann T, Kessler-Icekson G, et al. Isolation and characterization of the novel popeye gene family expressed in skeletal muscle and heart. Dev Biol 2000;223:371-82.
- Osler ME, Chang MS, Bader DM. Bves modulates epithelial integrity through an interaction at the tight junction. J Cell Sci 2005;118:4667-78.
- Smith TK, Hager HA, Francis R, et al. Bves directly interacts with GEFT, and controls cell shape and movement through regulation of Rac1/Cdc42 activity. Proc Natl Acad Sci U S A 2008;105:8298-303.
- Hager HA, Roberts RJ, Cross EE, et al. Identification of a novel Bves function: regulation of vesicular transport. EMBO J 2010;29:532-45.
- Williams CS, Zhang B, Smith JJ, et al. BVES regulates EMT in human corneal and colon cancer cells and is silenced via promoter methylation in human colorectal carcinoma. J Clin Invest 2011;121:4056-69.
- Luo D, Huang H, Lu ML, et al. Abnormal expression of adhesion protein Bves is associated with gastric cancer progression and poor survival. Pathol Oncol Res 2012;18:491-7.
- Feng Q, Hawes SE, Stern JE, et al. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev 2008;17:645-54.
- Salskov A, Hawes SE, Stern JE, et al. Hypermethylation of CCND2 May Reflect a Smoking-Induced Precancerous Change in the Lung. J Oncol 2011;2011:950140.
- Swaminathan PD, Purohit A, Hund TJ, et al. Calmodulin-dependent protein kinase II: linking heart failure and arrhythmias. Circ Res 2012;110:1661-77.


