
Variant panorama in 1,385 index patients and sensitivity of expanded next-generation sequencing panels in arrhythmogenic disorders
Introduction
Cardiomyopathies and channelopathies are relatively frequent conditions for which genetic testing is recommended in various guidelines (1,2). The genetics of primary electric disorders are caused by pathogenic variants in genes encoding ion channels and cardiomyopathies have been attributed to variants in genes including those of the sarcomere, the cytoskeleton, and desmosomes. The most widely recognized indication for genetic testing is to identify a causative variant and subsequently provide pre-symptomatic or predictive cascade screening of family members. This ensures adequate clinical surveillance of carriers and allows non-carriers to be discharged from clinical follow-up. Recent advances in DNA sequencing technologies have made it possible to investigate large numbers of disease genes simultaneously by next-generation sequencing (NGS), making variant analysis much faster and cheaper. However, analysis of large numbers of genes identifies a number of sequence variants of uncertain clinical significance (VUS or class 3 variants). A rigorous process of interpretation is necessary to avoid misclassification (3). Nevertheless, cardiologists and clinical geneticists are facing a major challenge in determining the clinical relevance of NGS results. Therefore, we analyze only recognized disease genes with substantial evidence of causality when offering routine genetic testing.
LQTS is the most common inherited channelopathy. Pathogenic variants have been identified in at least 17 genes that code for key cardiac ion channels, structural membrane scaffolding proteins and for cardiac channel interacting proteins. Approximately 75% of all pathogenic variants are found in KNCQ1, KCNH2 and SCN5A genes that are responsible for the LQTS subtypes 1–3. For these three subtypes, a phenotype-genotype correlation is evident. LQTS1 patients often exhibit syncopes during swimming. LQTS2 patients are most susceptible to arousal and emotional distress whereas arrhythmic events in LQTS3 occur during sleep.
BrS is associated with syncope and sudden cardiac death (SCD) resulting from polymorphic ventricular tachycardia (VT) and ventricular fibrillation (VF). Diagnosis of the condition requires the presence of a type 1 Brugada pattern on the ECG: coved ST-segment and J point elevation ≥0.2 mV, followed by a negative T wave. Brugada syndrome has been associated with variants in more than 20 genes. Only pathogenic variants in SCN5A are definitively disease causing; for all other genes only limited evidence exists (4).
CPVT was first described in children who experienced syncopes or died suddenly as a result of bidirectional or polymorphic VT and VF precipitated by sudden increases in sympathetic tones, such as physical exertion or emotional stress (5). The condition is highly penetrant and usually autosomal dominant inherited. Approximately 40–65% of probands carry a mutation in the RYR2 gene, which encodes the cardiac ryanodine receptor. Male carriers have a fourfold increased risk of cardiac events compared to female carriers. β-blocker therapy is, therefore, recommended even in asymptomatic RYR2-mutation carriers.
HCM is morphologically characterized by hypertrophy of the left ventricle. It is the most common form of the inherited cardiomyopathies with a prevalence of 1:500. HCM is primarily inherited in an autosomal pattern. HCM may manifest at any time from infancy to old age, but in the majority of patients symptoms do not set in before adolescence. About 50–60% of patients manifesting during adolescence or later harbor genetic defects in cardiac sarcomeric proteins (6).
DCM is characterized by dilation and limited contraction of the left or of both ventricles. The prevalence is approximately 1 in 2,500. The disease is usually accompanied by progressive cardiac insufficiency. DCM is generally detectable with echocardiography. There is a significant risk of arrhythmia and sudden cardiac death. Genetic testing including TTN has a diagnostic yield of up to 40% (7,8). The inheritance is mostly autosomal dominant.
ARVC is a mostly autosomal dominant inherited disorder of the heart muscle in which the myocardium is progressively replaced by fat and connective tissue. This fibrofatty remodeling mainly affects the right ventricle and leads to impaired conduction resulting in ventricular arrhythmias, palpitations or syncopes. ECG typically shows epsilon waves as well as an inverted T wave with broadened QRS complex in the right precordial leads. Around one third of index patients die suddenly at the age of 14–20. However, 50% of all carriers do not develop clinical symptoms until >50 years of age, and one third does not even present with symptoms in old age. Prevalence of ARVC is estimated to be 1:5,000–1:2,000. In 50–60% of patients, pathogenic variants can be detected in desmosomal genes (DSP, PKP2, and DSG2) (9).
Methods
Patients
Between 2016 and 2017, a total of 1,385 consecutive unrelated patients with a suspected diagnosis of LQTS, BrS, CPVT, HCM, DCM or ARVC were referred for genetic testing. Clinical data were provided for only 60% of the cases.
NGS
Genomic DNA was analyzed by NGS using a custom design based on an Agilent SureSelectQXT solution-capture enrichment strategy (Agilent, Santa Clara, CA, USA). Targeted sequencing capture probes were custom designed using the specific online tool (SureDesign) provided by Agilent in order to identify disease-causing variants in 83 genes (transcript variants were used according to HGMD® Professional): ABCC9, ACTC1, ACTN2, AKAP9, ANK2, ANKRD1, BAG3, CACNA1C, CACNA2D1, CACNB2, CALM1, CALM2, CALR3, CASQ2, CAV3, CRYAB, CSRP3, DES, DMD, DSC2, DSG2, DSP, FKTN, FLNC, GPD1L, HCN4, ILK, JPH2, JUP, KCND3, KCNE1, KCNE2, KCNE3, KCNE5, KCNH2, KCNJ2, KCNJ5, KCNJ8, KCNQ1, LAMA4, LAMP2, LDB3, LMNA, MYBPC3, MYH6, MYH7, MYL2, MYL3, MYLK2, MYOZ2, MYPN, NEBL, NEXN, PKP2, PLN, PRDM16, PRKAG2, RAF1, RANGRF, RBM20, RYR2, SCN10A, SCN1B, SCN2B, SCN3B, SCN4B, SCN5A, SGCD, SLMAP, SNTA1, TAZ, TCAP, TECRL, TGFB3, TMEM43, TNNC1, TNNI3, TNNT2, TPM1, TRDN, TRPM4, TTN, and VCL.
Target regions included coding exons (padding: _25 bp). The manufacturer’s protocols are followed during library preparation. For each sample, 50 ng of high quality genomic DNA was fragmented and adaptors were added in a single enzymatic step with QXT reagents. The adaptor-tagged DNA library was purified and amplified. The libraries were recovered using streptavidin magnetic beads, and a post-capture PCR amplification was carried out. Post-enrichment pooling allowed sequencing of a high variety of sample amounts as well as different sample target sizes. Sequencing was carried out on a Next-Seq500 sequencer using the NextSeq500 Mid or High Output Kit v2 (300 cycles) chemistry (Illumina, San Diego, California). Validation of enrichment and quantification of enriched target DNA were performed on Qubit Fluorometric Quantitation using Qubit dsDNA HS Assay kit (ThermoFisher Scientific, Illkirch, France) and the TapeStation 2,200 (Agilent). Fastq files (8 fastq files/patient) were generated using the Illumina provided tool bcl2fastq on an in-house server. Further bioinformatics analyses of small variants and copy number variants (CNVs) were further performed using CLC Genomics Workbench (Qiagen, Venlo, Netherlands) and a custom developed pipeline at the Center for Human Genetics. Copy number variants were detected with a combination of ExomeDepth, XHMM, and CoNVaDING. Identified putative pathogenic variants (SNV, indels) were verified by conventional dideoxy sequencing using BigDye Terminator v.3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, California), MLPA (MRC Holland) or qPCR. According to the recently reported guidelines, specific standard terminology (“pathogenic, class 5”, “likely pathogenic, class 4”, “uncertain significance, class 3”, “likely benign, class 2” and “benign, class 1”) were further used to evaluate the putative pathogenicity of variants identified in studied genes (3). Only class 3, 4 or 5 variants were taken into account in figures and online: http://fp.amegroups.cn/cms/0333ca39cab639893ad880e964e959c5/cdt.2019.06.06-1.pdf. Loss of function variants in the TTN-gene was classified as probably pathogenic (class 4) if at least the main transcript variants (NM_001267550 and NM_001256850) are affected and as class 3 if not. TTN missense variants, which are frequent in the general population, are not considered.
Statistical methods
The diagnostic yield or diagnostic sensitivity was calculated as proportion of class 4 or 5 positive patients/all patients with the same indication.
Results
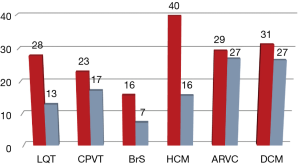
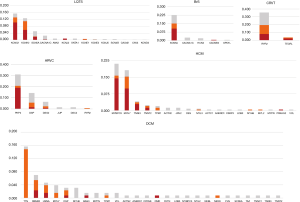
Genetic analysis was performed in 1,385 subjects with a suspected diagnosis of LQTS (649 cases), BrS [152], CPVT [88], HCM [242], DCM [128] or ARVC [126]. This unraveled the causal genetic defect in 397 (29%) of them. An additional 281 individuals (14%) had variants of uncertain significance (class 3). Pathogenic or likely pathogenic (class 5 or 4) variants were detected depending on indication in 16–40% of patients (Figure 1). Only five copy number variants showed up in 0.4% of the cases. About 1% of the patients carry more than one variant (data not shown). Diagnostic sensitivity was 28% for LQTS, 16% for BrS, 23% for CPVT, 29% for ARVC, 40% for HCM and 31% for DCM. As expected, 97% of the pathogenic variants were found in the well-known core genes. Only few genes substantially contribute to the diagnostic sensitivity (class 4 and 5 variants): for LQTS (26.1%) KCNQ1 (13.1%), KCNH2 (11.0%), and SCN5A (2.0%); for BrS SCN5A (15.1%); for CPVT RYR2 (19.5%); for HCM (36.7%) MYBPC3 (15.7%), MYH7 (15.3%), TNNI3 (3.7%), and TNNT2 (2.0%); for DCM (28.1%) TTN (14.8%), RBM20 (5.4%), LMNA (3.9%), DSP (2.4%), and MYH7 (1.6%) and for ARVC (28.6%) PKP2 (20.6%), DSP (5.6%), and DSG2 (2.4%) (Figure 2, http://fp.amegroups.cn/cms/0333ca39cab639893ad880e964e959c5/cdt.2019.06.06-1.pdf). Only a few additional genes account for a significant increase in diagnostic yield.
Interesting results showed in the analysis of the TECRL gene where patients with homozygous or combined heterozygous patients revealed a combined LQTS/CPVT phenotype. A 15-year-old son of consanguineous parents presented with a QTc of 520 ms and polymorphic ventricular tachycardia. He has a novel homozygous pathogenic variant c.415C > T, p.(Gln139*) which was transmitted from the heterozygous healthy parents. The second index patient suffered from syncopes, ventricular arrhythmias, bidirectional ventricular extrasystoles and showed a QTc of 450 ms. He has a pathogenic variant c.926C > A, p.(Ser309*) in combined heterozygosity with the probably pathogenic variant c.893T > C, p.(Val298Ala).
Moreover, pathogenic KCNJ2 variants were detected in 5 LQTS7 or Andersen-Tawil syndrome patients so that the analysis of KCNJ2 should always be included.
Several class 3 variants were detected in ANK2, KCNJ5, SCN4B (LQTS), CACNA1C (LQTS, BrS), HCN4 (BrS), ACTN2, JPH2, DES (HCM), JUP, DSC2 (ARVC), MYH6 and TTN (DCM). Genes with a remarkably high ratio of class 3 variants were ANK2, CACNA1C, SCN5A, MYH6, and TTN. Lots of class 3 variants were detected in DCM and ARVC patients (27%) in particular. One or two class 3 variants were found in most of the recently reported disease genes. Many patients have TTN missense variants which were not considered nor classified. One hundred and eighty-seven variants (38%) were probably not described so far as they were not listed in the HGMD or ClinVar databases. Sixteen of the twenty TTN class 4 loss of function variants described here were novel.
Discussion
We studied the genetic basis of arrhythmogenic disorders in 1,385 subjects referred to the Center for Human Genetics and Laboratory Diagnostics between 2016 and 2017. The major finding of this study was that the genetic cause of these could be solved in 29% of the patients. 14% of the patients had unclear reports caused by class 3 variants. As expected the vast majority of pathogenic variants (97%) were detected in the well-known 13 core genes KCNQ1, KCNH2, SCN5A, RYR2, MYBPC3, MYH7, TNNT2, TNNI2, LMNA, TTN, PKP2, DSP, and DSG2. The additive value of expanded panels is limited so that the analysis could be restricted to the core genes without significant loss of diagnostic yield. Only variants in these genes are highly actionable and interpretable in the clinic. Patients with more than one variant 1% (data not shown) were relatively rare in our collective compared to up to 5% in the literature. Only five copy number variants showed up in 0.4% of the cases. One hundred and eighty-seven variants were so far undescribed including sixteen TTN class 4 loss of function variants.
The diagnostic yield of 28% for LQTS was in the range of actual studies (30%) (10). The vast majority of the variants were detected in the core genes KCNQ1, KCNH2, and SCN5A. In patients with a suspected diagnosis of LQTS7 or Andersen-Tawil syndrome it is worth analyzing the KCNJ2 gene, and in patients with Timothy syndrome CACNA1C should be included. Additional interesting results were shown by the analysis of the TECRL gene where only three patients have been described so far (11). Homozygous or combined heterozygous patients revealed a combined LQTS/CPVT phenotype. For these patients flecainide could be used as a potential therapeutic. These findings have implications for diagnosis and treatment of inherited cardiac arrhythmias.
The diagnostic sensitivity for BrS of 16% was a little bit lower compared to other studies (20%) (12). Our results underline the recommendations to restrict the analysis to the only clinically valid gene SCN5A (4).
The analysis of the RYR2 gene revealed 23% causative variants in CPVT patients. This was rather low compared to other studies (60%, Roston et al., 2018, Europace 20:541). Low sensitivity is probably attributable to the fact that we have no inclusion criteria and many patients had few, unspecific or borderline clinical symptoms. Including class 3 variants with the possibly pathogenic variants as in older studies would raise diagnostic sensitivity to 40% for CPVT so that the range is comparable to other studies (5).
For HCM the diagnostic yield was 40% and an additional 16% showed inconclusive results. Both are in the range of other studies (13,14). The additive value of expanded panels is limited so that the analysis could be restricted to the core genes MYBPC3, MYH7, TNNI3, and TNNT2 without a significant loss of diagnostic yield. Only the TPM1 gene with 2.1% pathogenic variants should be added.
The analysis of our DCM gene panel results in a diagnostic yield of 31% which is comparable to the literature data (6,15,16). 28.1% of the variants were detected in the core genes TTN, RBM20, LMNA, DSP, and MYH7. The analysis of other newer DCM genes results in about 20% additional unclear reports. The identification of rare loss of function TTN variants could result in early-onset atrial fibrillation (17). Such truncating titin variants seem to be associated with a mild and treatable form of dilated cardiomyopathy (18).
Causative variants were detected in 29% of the ARVC patients. All these variants were located in the desmosomal core genes PKP2, DSP, DSG2 and one in RYR2. The diagnostic sensitivity is lower than in other studies 50–63% (19,20). A high proportion of the class 3 variants could not be avoided as a big part of these variants came up in the analysis of the core genes.
Conclusions
A definite genetic diagnosis can be established in 29% of patients with arrhythmogenic disorders. Genetic analysis was performed in 1,385 consecutive index patients with a suspected diagnosis of LQTS, BrS, CPVT, HCM, DCM or ARVC. The diagnostic sensitivity was lowest for BrS (16%) and highest for HCM (40%) and in the range of or lower than in the literature. The vast majority of pathogenic variants (97%) were detected in the 13 well-known core genes KCNQ1, KCNH2, SCN5A, RYR2, MYBPC3, MYH7, TNNT2, TNNI2, LMNA, TTN, PKP2, DSP, and DSG2. The additional inclusion of many genes could not significantly increase the diagnostic sensitivity. The additive value of expanded panels is limited so that the analysis could be restricted to the core genes. Only variants in these genes are highly actionable and interpretable in the clinic. The analysis of KCNJ2 (LQTS, Andersen-Tawil syndrome), RBM20 (DCM), TPM1 (HCM), and TECRL (LQTS/CPVT) could resolve the genetic basis of disease in some families and these genes should always be included.
Acknowledgments
We thank the patients and their families for the participation in the study. A consortium consisting of the Center for Human Genetics and Laboratory Diagnostics Dr. Klein, Dr. Rost and Colleagues, Genomatix GmbH, kbo—Kliniken des Bezirks Oberbayern and IMGM Laboratories was founded to develop the MIDAS software system for data integration in diagnostics. The MIDAS project is supported by a grant of the Bavarian Ministry of Economics Affairs and Media, Energy and Technology (Grant Number: “MED-1603-0011”).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Elliott PM, Anastasakis A, Borger MA, et al. ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy. Eur Heart J 2014;35:2733-79. [PubMed]
- Charron P, Arad M, Arbustini E, et al. Genetic counselling and testing in cardiomyopathies: a position statement of the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 2010;31:2715-26. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Hosseini SM, Kim R, Udupa S, et al. Reappraisal of Reported Genes for Sudden Arrhythmic Death. Circulation 2018;138:1195-205. [Crossref] [PubMed]
- Priori SG, Napolitano C. Cardiac and skeletal muscle disorders caused by mutations in the intracellular Ca2+ release channels. J Clin Invest 2005;115:2033-8. [Crossref] [PubMed]
- Walsh R, Thomson KL, Ware JS, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2017;19:192-203. [Crossref] [PubMed]
- van Spaendonck-Zwarts KY, van Rijsingen IAW, van den Berg MP, et al. Genetic analysis in 418 index patients with idiopathic dilated cardiomyopathy: overview of 10 years’ experience. Eur J Heart Fail 2013;15:628-36. [Crossref] [PubMed]
- Herman DS, Lam L, Taylor MRG, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med 2012;366:619-28. [Crossref] [PubMed]
- Vimalanathan AK, Ehler E, Gehmlich K. Genetics of and pathogenic mechanisms in arrhythmogenic right ventricular cardiomyopathy. Biophys Rev 2018;10:973-82. [Crossref] [PubMed]
- Lieve KV, Williams L, Daly A, et al. Results of Genetic Testing in 855 Consecutive Unrelated Patients Referred for Long QT Syndrome in a Clinical Laboratory. Genet Test Mol Biomarkers 2013;17:553-61. [Crossref] [PubMed]
- Devalla HD, Gélinas R, Aburawi EH, et al. TECRL, a new life-threatening inherited arrhythmia gene associated with overlapping clinical features of both LQTS and CPVT. EMBO Mol Med 2016;8:1390-408. [Crossref] [PubMed]
- Di Resta C, Pietrelli A, Sala S, et al. High-throughput genetic characterization of a cohort of Brugada syndrome patients. Hum Mol Genet 2015;24:5828-35. [Crossref] [PubMed]
- Mazzarotto F, Girolami F, Boschi B, et al. Defining the diagnostic effectiveness of genes for inclusion in panels: the experience of two decades of genetic testing for hypertrophic cardiomyopathy at a single centeret. Genet Med 2019;21:284-92. [Crossref] [PubMed]
- Alfares AA, Kelly MA, McDermott G, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med 2015;17:880-8. [Crossref] [PubMed]
- Bondue A, Arbustini E, Bianco A, et al. Complex roads from genotype to phenotype in dilated cardiomyopathy: scientific update from the Working Group of Myocardial Function of the European Society of Cardiology. Cardiovasc Res 2018;114:1287-303. [Crossref] [PubMed]
- Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol 2013;10:531-47. [Crossref] [PubMed]
- Ahlberg G, Refsgaard L, Lundegaard PR, et al. Rare truncating variants in the sarcomeric protein titin associate with familial and early-onset atrial fibrillation. Nat Commun 2018;9:4316. [Crossref] [PubMed]
- Jansweijer JA, Nieuwhof K, Russo F, et al. Truncating titin mutations are associated with a mild and treatable form of dilated cardiomyopathy. Eur J Heart Fail 2017;19:512-21. [Crossref] [PubMed]
- Bao J, Wang J, Yao Y, et al. Correlation of Ventricular Arrhythmias With Genotype in Arrhythmogenic Right Ventricular Cardiomyopathy. Circ Cardiovasc Genet 2013;6:552-6. [Crossref] [PubMed]
- Kapplinger JD, Landstrom AP, Salisbury BA, et al. Distinguishing Arrhythmogenic Right Ventricular Cardiomyopathy/Dysplasia–Associated Mutations From Background Genetic Noise. J Am Coll Cardiol 2011;57:2317-27. [Crossref] [PubMed]


