
Insights into the genetic basis of HMGB1 in atrial fibrillation in a Chinese Han population
Introduction
Atrial fibrillation (AF) is the most common cardiac arrhythmia, and profoundly increased the mortality, morbidity and healthy costs worldwide (1). In the past, traditional risk factors for AF, such as age, gender, smoking, hypertension and diabetes, have been illustrated fully and made a major contribution in the prevention and treatment of the disease (2-5). However, paroxysmal AF in young and middle-aged athletes was discovered with no traditional risk factors, which might indicate other factors with genetic basis involved in AF (6). Though, genomewide association studies have identified at least 30 genetic risk loci for AF, and explored lots of new molecular mechanisms for the development of AF (7-14), the exact pathology of AF still remains largely to be explained. Therefore, other studies, such as candidate gene association analysis, might be an important way to investigate the genetic basis of AF.
High mobility group box 1 (HMGB1) protein is a multifunctional redox sensitive protein and act both as a nuclear factor and a secreted protein (15-17). In 2009, Hu et al. reported that the serum level of HMGB1 was markedly increased with the severity of coronary artery stenosis in the patients of stable angina pectoris, which indicated that HMGB1 might involve in the development of coronary artery diseases, and the latter is one of the major cause for AF (18). In 2010, the same team studied in rats that, HMGB1 might improve the myocardial ischemia by decreasing the cell viability and promoting the apoptosis of neonatal myocytes, of which the latter might increase the incidence of AF (19). In the same year, Giallauria et al. reported that the serum HMGB1 levels were highly correlated with the autonomic dysfunction expressed by post-exercise slower HRR in post-infarction patients, which might increase the incidence of adverse cardiovascular events, such as AF and VF (20). In 2011, another research team found that the serum level of HMGB1 in AF patients was much higher than that of the control subjects (21). In 2013, Wu et al. studied that HMGB1 might improve the development of AF by increasing the oxidative stress in the patients (22). These evidences above indicated that there was a strong relationship between HMGB1 and AF, but the exact relationship between them are not clear yet.
Therefore, we investigated the genetic basis of HMGB1 in AF: we selected the variants locus in the regulatory region of HMGB1, and studied the genetic association between the selected variants and AF in a Chinese Han population.
Methods
Study population
In total, 576 AF cases and 869 controls were selected from the People’s Hospital of Yichang Center (Hubei, China). AF cases were diagnosed by cardiologists according to the guidelines (23). The control subjects were selected from the individuals with normal electrocardiogram and have no history of AF or any other cardiac arrhythmias. All of the individuals with type 1 diabetes, congenital heart disease, and heavy renal or hepatic diseases were excluded from this study. The control subjects with rheumatic autoimmune disease, tumor or stroke were also excluded from this study. The clinical characteristics, such as age, gender, history of smoking status, hypertension and diabetes mellitus, were obtained by direct interviews and medical record reviews.
This study was approved by the Ethics Committee of People’s Hospital of Yichang Center, and conformed to the ethical principles set forth by the Declaration of Helsinki. The informed consent was obtained from all the participants.
SNP selection
We selected the tag variants as following rules: first, the minor allele frequency (MAF) of the SNPs was greater than 0.05; second, the variants were given priorities, if they were previously reported functional variants or predicted potential functional sites by bioinformatics (Promoter, Genevar) (24). Two SNPs, rs1045411T/C nor rs1412125C/T, were selected according to the rules above. Rs1412125C/T locus in the promoter region of HMGB1 and rs1045411T/C locus in the 3’UTR region of HMGB1, of which both might regulate the gene expression of HMGB1.
Genotyping
We extracted the DNA samples from the peripheral blood of the studied population, following the standard process of the kit (the Wizard Genomic DNA Purification Kit, Promega Corporation, Madison, WI). Genotyping was performed with the selected SNPs using a Rotor Gene 6000 High-Resolution Melt (HRM) system (Corbett Life Science, Concorde, NSW, Australia). The PCR reaction system was in a total of 25 µL PCR volume containing 1 µL of LC Green dye, 5 pmol of each primer, 25 ng of genomic DNA, 2.5 µL of 10× PCR buffer with 1.5 mmol/L MgCl2, 5 mmol deoxynucleotide triphosphates, and 1 unit of Taq polymerase. Genotyping results were confirmed by Sanger sequencing.
Statistical analysis
Hardy-Weinberg disequilibrium tests were carried out for the selected SNPs in the control subjects using PLINK software version 1.07. The allelic and genotypic association analysis, as well as the odds ratio (OR) and 95% confidence interval (CI), were performed by the conduction of Pearson’s chi-square tests of 2×2 or 2×3 contingency tables (v.21.0, SPSS, Inc., Chicago, IL, USA). Traditional risk factors, such as age, gender, the history of smoking, hypertension and diabetes mellitus, were adjusted as covariates using a logistic regression analysis (SPSS, v.21.0). The haplotypic analysis was performed using SPSS (v.21.0, Inc., Chicago, IL, USA).
Results
Population characteristics
The characteristics of the studied subjects were illustrated in Table 1. Subjects in the AF case group were much older than those in the control group. AF cases also showed higher prevalence of male percent, hypertension and diabetes mellitus. However, the prevalence of the smoking status, which is also an important traditional risk factor for AF, were not significantly different between the AF case group and the control group.
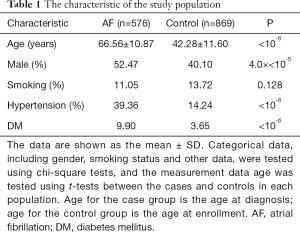
Full table
Association analysis between the selected SNPs of HMGB1 and AF in a Chinese Han population
Both of the two selected variants (rs1045411T/C nor rs1412125C/T) in HMGB1 passed the Hardy-Weinberg disequilibrium tests in the control subjects (P>0.05) (Table 2). Under the allelic association analysis, rs1045411T was not associated with AF neither before nor after the adjustment of the traditional risk factors in the Chinese Han population (Pobs=0.520, Padj=0.839, OR =1.03; 95% CI: 0.77–1.38); rs1412125C also showed no association with AF neither before nor after the traditional risk factors for AF in the Chinese Han population with all P values more than 0.1 (Pobs=0.982, Padj=0.715, OR =0.95; 95% CI: 0.73–1.24) (Table 2). Under the genotypic association analysis, rs1045411T/C showed a marginally significant association result with AF in the recessive model with adjusted p value of 0.056 (OR =0.42; 95% CI: 0.17–1.02); rs1412125C/T still showed no significant association with AF with all P values more than 0.1 in all the models (additive, dominant and recessive models) (Table 3). Under the haplotypic association analysis, we still found that the distribution frequency of the haplotypes had no significant difference between the AF cases and the controls with all P values more than 0.1 (Table 4).

Full table
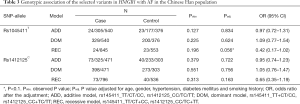
Full table

Full table
Association analysis between the selected variants and AF in the gender subgroup
We classified all the studied populations by gender and then investigated the association between the two SNPs (rs1045411T/C nor rs1412125C/T) and AF in different gender subgroups. Under the allelic association analysis, none of the two variants showed significant association with AF in the two gender subgroups with all P values more than 0.1 (Table 5). Under the genotypic association analysis, rs1045411T/C showed marginally significant association with AF in the male subgroup in recessive model (Pobs=0.065) (Table 5). However, after adjusted the traditional risk factors for AF, this association result did not remain with the adjusted P value of 0.104 (OR =0.36; 95% CI: 0.11–1.23) (Table 5). Rs1412125C/T again showed no association with AF under the genotypic association analysis neither in the male subgroup nor in the female subgroup in any models with all P values more than 0.1 (Table 5). Under the haplotypic association analysis, the combination type rs1045411T-rs1412125T was marginally associated with AF [(Padj=0.097, OR =0.67; 95% CI: 0.42–1.07) (Table S1); and rs1045411C-rs1412125C also showed marginally association with AF with an adjusted P value of 0.086 (OR =0.71; 95% CI: 0.48–1.05) (Table S1)].
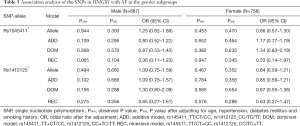
Full table
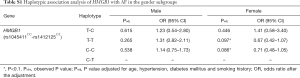
Full table
Association analysis between the selected variants and AF in the age onset subgroup
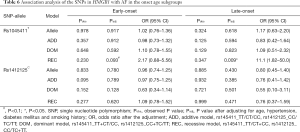
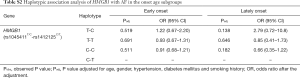
We divided the AF case population into two subgroups by the age onset of AF and investigated the association between the two SNPs (rs1045411T/C nor rs1412125C/T) and AF in different age onset subgroup. Patients with the age onset of AF less than 65 years were defined as the early-onset AF subgroup, and the other patients were defined as the late-onset AF subgroup. Under the allelic association analysis, no significant association results were found between the two SNPs and AF neither in the early-onset AF subgroup nor in the late-onset AF subgroup with all P values more than 0.1 (Table 6). Under the genotypic association analysis, rs1045411T showed marginally significant association with AF in the early-onset subgroup, and highly significant association with AF in the late-onset subgroup (early-onset, Padj=0.093, OR =0.46; 95% CI: 0.18–1.14; late-onset, Padj=0.009, OR =11.1; 95% CI: 1.82–50.0) (Table 6); while rs1412125C showed no significant association with AF neither in the early-onset subgroup nor in the late-onset subgroup. In addition, the haplotypic association analysis showed no significant association results neither in the early-onset subgroup, nor in the late-onset subgroup with all P values more than 0.1 (Table S2).
Full table
Full table
Discussion
In this study, we investigated the genetic basis of HMGB1 in AF in a Chinese Han population. In total study population, the promoter variant rs1412125C/T was not associated with AF neither under the allelic association analysis, nor under the genotypic association analysis; the 3’UTR variant rs1045411T/C in HMGB1 was not associated with AF under the allelic association analysis, but under the genotypic association analysis, rs1045411T was marginally associated with AF in the recessive model; the haplotypes of the two SNPs (rs1045411T/C nor rs1412125C/T) in HMGB1 were not associated with AF. In the gender subgroup, neither rs1412125C/T nor rs1045411T/C showed significant association with AF under the allelic or genotypic association analysis; the combination types of rs1045411T-rs1412125T and rs1045411C-rs1412125C showed marginally association with AF under the haplotypic association analysis in the female subgroup. In the age onset subgroups, rs1412125C was not associated with AF in any subgroup; rs1045411T was marginally associated with early onset AF and significantly associated with late-onset AF under the genotypic association analysis in the recessive model; the haplotypic association analysis showed none significant association results in any subgroups.
HMGB1 is a highly conserved chromatin protein and binds with DNA and regulates other genes’ expression in the nucleus. It can also act as proinflammatory cytokine, which could be released by necrotic or damaged cells and activated immune cells, such as macrophages and monocytes (15,25). Researchers have reported that the serum levels of HMGB1 were increased much higher in the patients with inflammatory diseases, such as endotoxemia, sepsis, reperfusion injury, acute lung injury or autoimmune disease, myocardial ischemia, acute coronary syndrome and AF (15,18,19,21,26-28). In 2012, Hu et al. review that the HMGB1 might participate in the development of AF, by promoting oxidative stress, matrix metalloproteinase-9 upregulation and activation or the atrial structural remodeling (18-21,29). In 2015, Qu et al. reported that the intron variant of HMGB1, rs2249825, was associated with postoperative AF after coronary artery bypass grafting (CABG) under cardiopulmonary bypass (CPB) in a Chinese Han population (30). In above, HMGB1 might play an important role in the development of AF and might be an important candidate gene for the risk of AF.
In this study, we selected two common variants located in the regulatory region of HMGB1, the promoter variant rs1412125C/T and the 3’UTR variant rs1045411T/C, to test the association between the two variants in HMGB1 and AF in a Chinese Han population. In 2016, Bao et al. first confirmed that the variant rs1045411, which is in close proximity to the microRNA (hsa-miR-505) binding site in the 3’UTR region of HMGB1 gene, could significantly influence the mRNA expression of HMGB1 in gastric cancer tissues and two tumor cell lines (SGC-7901 and HEK-293T cell lines) (31-33). In 2017, Lin et al. also determined that the 3’-UTR SNP rs1045411T/C could alter the mRNA stability of HMGB1 and then increase risk to squamous cell carcinoma (OSCC) (34). In above, we might conclude that rs1045411T/C might be an important expression quantitative trait loci (e-QTL) for HMGB1. In recent years, lots of researchers discovered that the e-QTLs, which were variants regulated the expression of the genes, had the population heterogeneity, the tissue cell specificity and influence the risk of disease under different conditions (35,36). In our study, we found that rs1045411T/C significantly contributed to the risk of late onset AF, which indicated that rs1045411T/C might just be the e-QTL for HMGB1 in the old patients.
In 2014, Wang et al. reported that rs1412125C/T and rs2249825G/C in HMGB1 were associated with platinum-based chemotherapy responses in Chinese lung cancer patients, which indicated that the two variants (rs1412125C/T and rs2249825G/C) in HMGB1 might be important biomarkers for predicting the efficacy of platinum-based chemotherapy (37). In 2016, Wu et al. discovered that rs1412125C/T and rs2249825G/C in HMGB1 were associated with susceptible to the development of cervical invasive cancer in Taiwanese Women in the Chinese population (38); another team reported that the promoter variant rs1412125C/T in HMGB1 was associated with hepatocellular carcinoma (HCC) in Taiwan, which indicated that rs1412125C/T might be a genetic risk factor for HCC in the Chinese population (39). Here, we didn’t find the association between the promoter variant rs1412125C/T in HMGB1 and AF, which indicated that rs1412125C/T was not a genetic risk factor for AF. In addition, we didn’t select the variant rs2249825G/C because of its locus in the intron region of HMGB1, which might have no function in regulating of HMGB1 expression and might not be a functional risk variant for any diseases.
Conclusions
Here, we might conclude that HMGB1 might be an important causal factor for the development of AF, and rs1412125C/T might be an important genetic risk factor for AF. All of these evidences provide important information in the study of treatment and prevention for AF. Further studies with larger sample size, functional study of HMGB1 in older subjects and interaction with other genes or microRNAs are needed to confirm our results.
Acknowledgments
Thanks to all the participants.
Funding: This work was supported by grants from National Natural Science Foundation of China [No. 81400246 to L Li].
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cdt.2019.12.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of People’s Hospital of Yichang Center, and conformed to the ethical principles set forth by the Declaration of Helsinki. The informed consent was obtained from all the participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Weng LC, Preis SR, Hulme OL, et al. Genetic Predisposition, Clinical Risk Factor Burden, and Lifetime Risk of Atrial Fibrillation. Circulation 2018;137:1027-38. [Crossref] [PubMed]
- Benjamin EJ, Levy D, Vaziri SM, et al. Independent risk factors for atrial fibrillation in a population-based cohort: the Framingham Heart Study. JAMA 1994;271:840-4. [Crossref] [PubMed]
- Wang TJ, Parise H, Levy D, et al. Obesity and the risk of new-onset atrial fibrillation. JAMA 2004;292:2471-7. [Crossref] [PubMed]
- Dublin S, French B, Glazer NL, et al. Risk of new-onset atrial fibrillation in relation to body mass index. Arch Intern Med 2006;166:2322-8. [Crossref] [PubMed]
- Thomas MC, Dublin S, Kaplan RC, et al. Blood pressure control and risk of incident atrial fibrillation. Am J Hypertens 2008;21:1111-6. [Crossref] [PubMed]
- Cervellin G, Sanchis-Gomar F, Filice I, et al. Paroxysmal atrial fibrillation in young and middle-aged athletes (PAFIYAMA) syndrome in the real world: a paradigmatic case report. Cardiovasc Diagn Ther 2018;8:176-9. [Crossref] [PubMed]
- Bapat A, Anderson CD, Ellinor PT, et al. Genomic basis of atrial fibrillation. Heart 2018;104:201-6. [Crossref] [PubMed]
- Sinner MF, Tucker NR, Lunetta KL, et al. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation 2014;130:1225-35. [Crossref] [PubMed]
- Gudbjartsson DF, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet 2009;41:876-8. [Crossref] [PubMed]
- Ellinor PT, Lunetta KL, Albert CM, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet 2012;44:670-5. [Crossref] [PubMed]
- Christophersen IE, Rienstra M, Roselli C, et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet 2017;49:946-52. [Crossref] [PubMed]
- Lee JY, Kim TH, Yang PS, et al. Korean atrial fibrillation network genome-wide association study for early-onset atrial fibrillation identifies novel susceptibility loci. Eur Heart J 2017;38:2586-94. [Crossref] [PubMed]
- Low SK, Takahashi A, Ebana Y, et al. Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat Genet 2017;49:953-8. [Crossref] [PubMed]
- Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007;448:353-7. [Crossref] [PubMed]
- Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002;418:191-5. [Crossref] [PubMed]
- Fink MP. Bench-to-bedside review: High-mobility group box 1 and critical illness. Crit Care 2007;11:229. [Crossref] [PubMed]
- Bell CW, Jiang W, Reich CF 3rd, et al. The extracellular release of HMGB1 during apoptotic cell death. Am J Physiol Cell Physiol 2006;291:C1318-25. [Crossref] [PubMed]
- Hu X, Jiang H, Bai Q, et al. Increased serum HMGB1 is related to the severity of coronary artery stenosis. Clin Chim Acta 2009;406:139-42. [Crossref] [PubMed]
- Hu X, Zhou X, He B, et al. Minocycline protects against myocardial ischemia and reperfusion injury by inhibiting high mobility group box 1 protein in rats. Eur J Pharmacol 2010;638:84-9. [Crossref] [PubMed]
- Giallauria F, Cirillo P, Lucci R, et al. Autonomic dysfunction is associated with high mobility group box-1 levels in patients after acute myocardial infarction. Atherosclerosis 2010;208:280-4. [Crossref] [PubMed]
- Hu XR, Zhou WJ, Bai QJ, et al. Increased serum high mobility group box 1 protein in patients with atrial fibrillation. Biomed Aging Pathol 2011;1:52-5. [Crossref]
- Wu Y, Zhang K, Zhao L, et al. Increased serum HMGB1 is related to oxidative stress in patients with atrial fibrillation. J Int Med Res 2013;41:1796-802. [Crossref] [PubMed]
- January CT, Wann LS, Alpert JS, et al. 2014 AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. J Am Coll Cardiol 2014;64:e1-76. [Crossref] [PubMed]
- Tu X, Nie S, Liao Y, et al. The IL-33-ST2L pathway is associated with coronary artery disease in a Chinese Han population. Am J Hum Genet 2013;93:652-60. [Crossref] [PubMed]
- Dumitriu IE, Baruah P, Manfredi AA, et al. HMGB1: guiding immunity from within Trends Immunol 2005;26:381-7. [J]. [Crossref] [PubMed]
- Wang H, Bloom O, Zhang M, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science 1999;285:248-51. [Crossref] [PubMed]
- Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol 2005;5:331-42. [Crossref] [PubMed]
- Andrassy M, Volz HC, Igwe JC, et al. High-mobility group box-1 in ischemia-reperfusion injury of the heart. Circulation 2008;117:3216-26. [Crossref] [PubMed]
- Hu XR, Wang XH, Liu HF, et al. High mobility group box 1 protein: possible pathogenic link to atrial fibrillation. Chin Med J (Engl) 2012;125:2346-8. [PubMed]
- Qu C, Wang XW, Huang C, et al. High mobility group box 1 gene polymorphism is associated with the risk of postoperative atrial fibrillation after coronary artery bypass surgery. J Cardiothorac Surg 2015;10:88. [Crossref] [PubMed]
- Betel D, Wilson M, Gabow A, et al. The microRNA.org resource: targets and expression. Nucleic Acids Res 2008;36:D149-53. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Bao G, Qu F, He L, et al. Prognostic Significance of Tag SNP rs1045411 in HMGB1 of the Aggressive Gastric Cancer in a Chinese Population. PLoS One 2016;11:e0154378. [Crossref] [PubMed]
- Lin CW, Chou YE, Yeh CM, et al. A functional variant at the miRNA binding site in HMGB1 gene is associated with risk of oral squamous cell carcinoma. Oncotarget 2017;8:34630-42. [Crossref] [PubMed]
- Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013;45:1238-43. [Crossref] [PubMed]
- Li X, Hastie AT, Hawkins GA, et al. eQTL of bronchial epithelial cells and bronchial alveolar lavage deciphers GWAS-identified asthma genes. Allergy 2015;70:1309-18. [Crossref] [PubMed]
- Wang Y, Li XP, Yin JY, et al. Association of HMGB1 and HMGB2 genetic polymorphisms with lung cancer chemotherapy response. Clin Exp Pharmacol Physiol 2014;41:408-15. [Crossref] [PubMed]
- Wu HH, Liu YF, Yang SF, et al. Association of single-nucleotide polymorphisms of high-mobility group box 1 with susceptibility and clinicopathological characteristics of uterine cervical neoplasia in Taiwanese women. Tumour Biol 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Wang B, Yeh CB, Lein MY, et al. Effects of HMGB1 Polymorphisms on the Susceptibility and Progression of Hepatocellular Carcinoma. Int J Med Sci 2016;13:304-9. [Crossref] [PubMed]


