
Cardiac multi-scale investigation of the right and left ventricle ex vivo: a review
Introduction
The heart, as other organs within the body, can be understood as a complex multi-scale system composed of components integrated at subcellular, cellular, tissue and organ levels, which work efficiently together in order to receive blood and subsequently pump it to the rest of the body. Cardiovascular diseases (CVDs), the leading cause of mortality in developed countries, can lead to cardiovascular remodeling affecting cardiac structure and function at all scales, from the sarcomere, the basic contractile unit within the myocyte, to the whole heart.
The cardiac myocytes, known also as cardiomyocytes, are the primary functional muscle cells in the myocardium (1). They are aggregated together to form a complex three-dimensional (3D) network showing predominant directions within the ventricular myocardium (2). Several studies have suggested that myocytes are also grouped forming flattened structures of approximately four myocytes thick although there is some controversy about the presence or absence of these structures. Different terms such as: ‘sheets’, ‘sheetlets’, ‘laminar’ or ‘lamellar’ have been used interchangeably to describe them (3,4). The orientation of myocyte aggregates is assessed by means of three different angles. Their longitudinal orientation with respect to the long axis of the ventricular cavity is known as helical angle; their radial orientation with respect to the epicardial surface is known as intrusion angle (also known as transverse, transmural or imbrication angle). Finally, the angle between the normal vector of the plane of myocytes aggregates and the epicardial surfaces is known as ‘sheet’-normal or E3 angle. There is continuing debate about precise structure, in particular on how these groups of myocytes are packed within their surrounding connective tissue matrix (the endo-, peri- and epi-mysium), and how they achieve this form in-utero (5,6). It is also known that myocardial architecture of the heart differs between the left ventricle (LV) and the right ventricle (RV). While the LV has a thick compact myocardium, the RV has a very complex shape and more trabeculations, especially in the apical part, with a thin compact myocardium. Unlike the LV, the normal RV myocardium is not thought to contain a mid-layer of circumferentially arranged myocytes apart from the outflow tract (7,8). Nonetheless, the arrangement of the myocytes within the ventricular walls determines propagation of the electrical activity through the myocardium as well as the development of force during cardiac contraction or relaxation (9,10). To understand cardiac dysfunction associated with CVDs, therefore, it is important to study the alterations that occur at all scales of the heart, from subcellular to whole organ level. This requires multi-scale imaging approaches that allow quantitative assessment of ventricular architecture and its remodeling ideally both ex vivo and in vivo.
Cardiovascular imaging is used widely in basic and clinical applications to investigate cardiac structure and function as well as for the diagnosis and assessment of cardiac remodeling in CVDs. Ventricular architecture has been extensively studied using microscopy by multiple technologies (light, confocal or electron microscopy) (11,12), which can be very powerful in terms of resolution and contrast thanks to histological staining agents, but involves destruction of the sample and only 2D images from thin tissue sections can be obtained. On the other hand, 3D imaging techniques such as ultrasound (US), magnetic resonance imaging (MRI) or (micro-) computed tomography (CT), amongst others, have proven popular in cardiology since they are able to image the whole heart both ex vivo and in vivo albeit with lower resolution than using microscopy (see Figure 1). Recently, synchrotron X-ray phase contrast imaging (SR X-PCI) has emerged as a powerful imaging technique for the investigation of cardiac structure ex vivo. This technique uses the refractive properties of brilliant X-rays to enhance image contrast in tomographic measurements, which is limited for soft tissues in conventional clinical systems. Thanks to its versatile sample stage and detector configuration, X-PCI offers the possibility to perform multi-resolution studies without sample manipulation (13), and with resolutions down to the submicron level (13,14-19). However, up to now, it is only possible to perform ex vivo studies in small samples (<3 cm3).
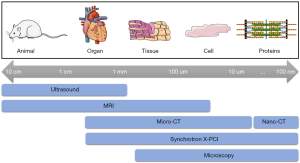
In this review, we provide a thorough overview of the imaging techniques that are currently being used to assess ventricular architecture at different scales, both in health and in disease, focusing on those than can provide details at different scales from the same sample. Table 1 provides a summary of the main characteristics of the different imaging modalities discussed in this review. We conclude with the future of research and challenges of multi-scale cardiovascular imaging.
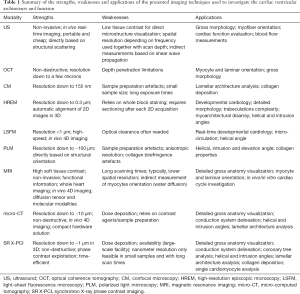
Full table
Ultrasound
Cardiac US, also known as echocardiography, is a non-invasive imaging modality, routinely used in clinical practice, that can provide dynamic cardiac anatomical and hemodynamic information thanks to its real-time nature. Almost 10 years ago, Lee et al. proposed the use of shear waves (a type of elastic wave) generated by US, travelling in different directions within the myocardium, to map the orientation of myocyte aggregates (20). This technique is known as shear wave imaging (SWI) or elastic tensor imaging (ETI). Since shear wave propagates faster along than across the myocyte direction, the helical angle of myocyte aggregates can be estimated by finding the maximum shear wave in vivo speed. Lee and colleagues successfully mapped the transmural fiber orientation in 5 in vitro porcine and 3 open-chest ovine hearts with a spatial resolution of 0.2 mm (20). In the in vitro porcine myocardium, the estimated helical angle of myocytes gradually changed from +80º (endocardium) to –40º (epicardium) with 0º aligning with the circumference of the heart, while in the in vivo hearts, the average orientation exhibited +71º (endocardium) and –26º (epicardium) at mid-systole. However, an important limitation of SWI is the need to generate a shear wave at every location of interest, thus requiring long acquisitions.
More recently, Papadacci et al. introduced 3D backscatter tensor imaging (3D-BTI) to map myocyte orientation dynamically with high temporal resolution in vivo (21). In 3D-BTI, ultrafast volumetric US acquisitions are used to measure the spatial coherence of ultrasonic speckle. The authors estimated myocyte orientation in vitro in 5 porcine LV myocardial samples and subsequent histology was performed in these samples for validation. In the same study the authors also reported the use of 3D-BTI in vivo, to assess the changes in myocyte orientation in the beating heart of an open-chest sheep as well as on a healthy volunteer to test its clinical feasibility. However, an important limitation of 3D-BTI is its incapability to map the z component of myocyte orientation as well as its low spatial resolution. Overall, although US is a cheap and non-invasive imaging technique that offers excellent temporal resolution for assessing global cardiac structure and function real-time it has a poor spatial resolution (few mm) to study myocardial architecture in detail.
Optical coherence tomography
Optical coherence tomography (OCT) is a non-destructive technique that utilizes light in the near-infrared spectral range to provide 3D images at micrometer-resolution from biological samples at high acquisition speeds (22).
OCT was first used to extract the 3D orientation of myocytic aggregates (23). Gan et al. acquired 3D images from swine hearts, without optical clearing and with a pixel size of about 5 µm, to quantify myocyte orientation. More recently, OCT has been used to image entire young and adult mouse hearts ex vivo at micron scale resolution, to investigate morphological changes in cardiac architecture that occur with normal aging (24), showing that the slope of myocyte orientation from the endocardium to the epicardium decreased with age. However, one of the main drawbacks of OCT is its limited depth of light penetration (about 1 mm) which restricts its use to small animals.
Polarization-sensitive optical coherence tomography (PS-OCT) has been used to assess myocyte orientation in fixed (25) as well as in a fresh mouse hearts (26). This technique is an extension of OCT which can determine depth-resolved local polarization properties. While conventional ‘standard’ OCT is based exclusively on the intensity of light backscattered or reflected by the sample, PS-OCT also detects its polarization thereby improving image contrast as well as allowing quantification of sample specific polarization properties. The authors showed that, although the imaging depth of the PS-OCT system was not able to penetrate the entire ventricular wall in mice, it was sufficient to quantify myocytes and sheet orientation in 3D and to validate this technique with histological measurements (26).
Microscopy imaging
Microscopy techniques are based on the interaction of sample sections with visible light with the aid of conventional histological staining or the use of antibody specific fluorescent or non-fluorescent probes. The high resolution achievable and the wide offer of contrast agents and biomarkers make microscopic techniques very powerful for the detailed study of biological tissues at (sub)cellular level. Nevertheless, these techniques introduce well-characterized alterations due to tissue processing, staining, embedding and cutting, and typically rely on the acquisition of individual 2D images from 3D samples. Among all the different microscopy techniques available, the ones that have been widely used to characterize ventricular architecture include; confocal, high-resolution episcopic microscopy (HREM), polarized light microscopy (PLM) and, more recently, light-sheet fluorescence microscopy (LSFM).
Confocal microscopy (CM)
CM is a scanning variation of traditional fluorescence microscopy that uses point illumination and a pinhole in front of the detector to eliminate out-of-focus signal. Therefore, only the fluorescence signal coming from the region close to the focal plane is detected and resolution is increased significantly (27). Thanks to staining procedures and hardware developments, high resolution images (<1 µm) of cardiac subcellular structures over relatively small volumes (up to 4 mm3) and areas (up to 75 mm2) can be imaged (28).
This technique, in combination with a variety of image processing tools, has been used to study cardiomyocyte orientation in chicken embryos with altered mechanical load (29), in fetal rabbits (30) and in rats (31,32). In addition, CM has been extensively used to investigate the fibrous matrix/collagenous microstructure and lamellar structure in adult rats (28,32,33), as well as its age-dependent remodeling in rat models of hypertension and its relationship with cardiac left ventricular function (34-36).
Although CM offers resolution and image contrast adequate to resolve cardiac tissue microstructural components ex vivo, a general limitation of this technique is related to the dehydration of tissue and use of chemicals which might alter inherent myocardial structure. Moreover, the visualization of cardiac microstructure of whole hearts with CM is not feasible, therefore limiting it to the investigation of small samples of cardiac tissue.
High-resolution episcopic microscopy
In HREM, high-resolution 2D digital images of the surface of resin-embedded samples, with pixel sizes ranging from 0.5 to 3 µm2, are acquired in fluorescence-mode after repeated sectioning. In contrast to traditional histology, this process is done automatically so that a perfectly aligned 3D dataset can be generated without user interaction, thus avoiding related manipulation distortions and enabling to decrease section thickness up to 1 µm (37). For a detailed review about HREM, we refer the reader to the publication of Weninger et al. (38).
Given its high resolution and the ability to generate 3D volumes, HREM has been extensively used to visualize cardiac morphology at different stages of fetal development in animal models such as zebrafish, frog, chicken and mouse (38-40) as well as in human (41,42). Among all the different animal models, the mouse has been most widely studied (43). While some studies focus on morphological development of normal cardiac structures (44-46), others have assessed defects induced by a variety of genetic mutations (40,47,48), thus showing the potential of HREM for high throughput phenotyping of mouse mutants.
Most of the literature is focused on visual inspection/3D reconstructions of cardiac morphology and only few performed quantitative assessment of myocardial architecture. The fractal dimension in combination with the measurement of ventricular thickness, size and surface has been proposed to quantify the complexity and development of trabeculations both in normal and in mutant mouse models (49-51). More recently, Garcia-Canadilla et al. quantified the orientation of myocytes aggregates (helical and intrusion angles) and myoarchitectural disarray in a mutant mouse model of hypertrophic cardiomyopathy during fetal development and at birth (52).
One of the limitations of HREM is that it is restricted to exploring the microanatomy of small specimens. Moreover, as for most microscopy techniques, it involves the dehydration and processing of samples. Finally, use of histologic stains and antibodies is still experimental and requires careful collection of all tissue sections obtained during the HREM process (40). Therefore, with this technique is still not possible to visualize specific cardiac tissue components such as the fibrous matrix and relate these to myocyte organization.
Polarized light microscopy
PLM is a microscopy technique that uses polarized light to image specimens that are visible primarily due to their optically anisotropic character (53). Generally, the setup consists of a first polarizer that polarizes the light coming from a white light source, next to the sample, and a second polarizer (analyzer) perpendicular to the first one. A charge-coupled device (CCD) camera, placed behind, detects the transmitted light. When the light interacts with an anisotropic sample, as in the case of cardiac tissue, its polarization axis changes and some of the light propagates through the analyzer. The intensity detected by the CCD camera is related to the main orientation axis of the sample.
Within the cardiovascular field, PLM has been mainly used for the visualization of myocardial collagen and quantification of its orientation both in animal models (54) and humans (55,56). Whittaker and colleagues proposed the use of circularly instead of linearly polarized light in combination with picrosirius red staining to enhance histological assessment of myocardial collagen thus providing additional insight into its composition and structure (54).
Moreover, PLM has been also used to quantitatively measure the orientation of myocyte aggregates in human fetal and infantile hearts embedded in methyl methacrylate by inferring the mean spatial orientation of the myosin filaments with a resolution of few microns, and described by means of the helical and intrusion angles (57-59). However, one of the main limitations is that the range of helical angle that can be measured is restricted to 90º having also a systemic bias between 0º and 20º. Moreover, values of helical angle higher than 70º cannot be resolved due to the low amount of transmitted light. Therefore, PLM seems to be more suitable for the quantitative analysis of myocardial collagen rather than the assessment of myocytes aggregates orientation, although it is restricted to the study of small pieces of tissue and also the intensity of the transmitted light depends on the orientation of collagen fibers with respect to the section plane.
Light-sheet fluorescence microscopy
LSFM is a non-destructive technique in which a thin sheet of light illuminates the sample and a fluorescence signal is detected orthogonally by a 2D detector, usually a CCD or complementary metal-oxide semiconductor (CMOS), combined with an objective (60). With the use of very thin light-sheets, deep-tissue penetration, high spatial resolution (<1 µm) and good optical sectioning with minimal photobleaching and phototoxicity is achieved, with an additional increase of contrast due to the reduction of background signal. The high spatiotemporal resolution offered by LSFM has been mainly used to study real-time developmental biology (61). The fast acquisitions achieved with respect to other microscopy techniques, allows in vivo visualization (4D imaging) of samples ranging from live zebrafish embryos to adult mouse hearts (62-64).
4D imaging of zebrafish embryos has been used to dynamically investigate cardiac anatomy, architecture and function (65,66) as well as for elucidating the mechanisms of cardiovascular regeneration and chemotherapy-induced cardiac remodeling (62,67). Some of these studies focused also on the detailed visualization of neonatal murine hearts, proving the capabilities of LSFM to unravel functional and structural information of the murine heart (64,66). However, all the studies found in the literature focused only on the visual inspection of the heart without providing any quantitative data about cardiac architecture and/or function.
Compared to other microscopy techniques such as CM or two-photon microscopy, LSFM is able to image thicker tissues (>1 cm) with reduced photobleaching and phototoxicity. Moreover, LSFM offers higher resolution and faster acquisitions compared with other non-destructive techniques such as MRI or CT. However, one of the main limitations is that since LSFM is an optical method samples need to be transparent and fluorescent for imaging, meaning that many samples require optical clearing in order to eliminate self-absorption of tissue and use of fluorophores.
Magnetic resonance imaging
MRI and, more specifically, diffusion tensor imaging (DTI) has become the gold standard for the investigation of myocardial architecture, both ex vivo and in vivo, thanks to the high anisotropy of the myocardium and the ability of DTI to measure water diffusion paths (68,69). DTI enables estimation of the three orthogonal eigenvectors of the diffusion tensor, v1, v2 and v3, that are associated to myocytes aggregates longitudinal, ‘sheet’ and ‘sheet’-normal directions respectively.
MRI offers superior soft tissue contrast in 3D without the use of ionizing radiation and thus the possibility to perform in vivo studies. Nevertheless, some authors have reported the use of gadolinium to increase contrast in 3D fast low angle shot MRI (FLASH) images or visualize myocardial fibrosis (70,71). Spatial resolution is typically limited (~1 mm), requiring long acquisition times (>12 h) for high resolution scans. For a review on the technical details of cardiac MRI we refer the reader to the works of Hoffman et al. (72) and Mekkaoui et al. (68).
An extensive volume of research supports DTI in characterizing myocyte aggregate orientation ex vivo, in experimental animal models such as mouse (73-75), rat (70,76-84), rabbit (71,85), bovine (86), ovine (87,88), canine (89-91), porcine (92-96) and even in human fetal (97) and adult (98,99) hearts. Most of these studies focused on the visualization and quantification of myocyte aggregate orientation in healthy hearts by means of helical, intrusion and sheet angles, trying to shed more light on the controversy about the precise myocardial architecture and the existence, or not, of a unique ventricular myocardial band (6,7). Furthermore, some studies have also quantified myocardial remodeling induced by different CVDs such as myocardial infarction (75,79) or pulmonary hypertension (88). Among all the different ex vivo MRI studies published over the last 20 years, that of Gilbert and colleagues demonstrated the highest resolution, with an isotropic voxel size of 50 µm3 (82).
Most of the ex vivo MRI studies discussed above focused only on describing myocardial architecture of the LV, while very few have investigated also RV myocardial architecture in detail, both in healthy conditions comparing different contraction states (71,94) and in different CVDs (88,100), which is crucial for a full understanding of the overall cardiac structure-function relationship. Considering that the RV wall is very thin (2–5 mm in humans), the limited spatial resolution offered by MRI could have been a limiting factor for investigating the myocardial architecture of RV.
Another topic of interest has been the study of the changes in myocyte orientation during cardiac contraction. This has been produced either by fixing hearts at different selected phases of the cardiac cycle and imaging them ex vivo (71,94), or by using in vitro isolated heart perfusion systems in rats (76,80,83), which allow to stop the same heart in different contraction states thanks to the use of different ion concentrations in the buffer solutions. Surprisingly, the different studies have reported different results. While Chen et al. have observed an increase in longitudinally arranged myocytes in both the endocardium and epicardium at the end of systole (80), Hales et al., and Teh et al., also reported an increase in the longitudinally arranged myocytes in the sub-endocardium but a decrease in longitudinally arranged myocytes in the sub-epicardium during contracture state (71,83). The authors attribute these differences to the chemical fixation of the heart in the contracture heart prior to imaging. More recently, Omann and colleagues have reported a decrease in E3-angle from relaxation to contraction, predominantly in the endocardium of both ventricles of female swine, with no significant changes in either the helical or intrusion angle of myocytes of the RV. These results suggest that the aggregation of myocytes is the predominant mediator of myocardial wall thickening during cardiac contraction (94).
Generally, in ex vivo MRI studies with long acquisition times, the sample is chemically fixed and embedded in either agarose or Fomblin to prevent tissue autolysis and sample motion. However, this may alter tissue structure and MRI properties as demonstrated by Hales et al. (81). The authors reported changes in T1 and T2 relaxation times over 48-h period following embedding in all the different media as well as alterations in the orientation of the primary eigenvector of the diffusion tensor in all the hearts. Therefore, the use of some chemicals for tissue fixation and embedding may alter tissue properties and this has to be taken into account when assessing changes in cardiac microstructure due to CVDs.
One of the great advantages of MRI is the possibility to perform in vivo studies non-invasively. This way, changes in myocyte and ‘sheet’ orientation during the cardiac cycle have been studied in vivo in healthy humans (101) as well as in patients with hypertrophic cardiomyopathy (102) and dilated cardiomyopathy (95), and further compared to in vivo porcine measurements. One of the main limitations of all these in vivo studies is the low spatial as well as temporal resolution. Furthermore, information was only acquired at specific short-axis slices of the heart thus limiting the possibility to detect small local changes in myocardial architecture. This could explain why these studies have showed limited changes in myocytes helical angle between different contractile states.
Most of the studies have tried to validate DTI using conventional histology, which requires destructive sectioning to acquire 2D optical images and which thus might alter myocyte microstructure, limiting the correct assessment of 3D myocardial architecture (77,78,82,83,85,86,95). Recently, DTI has been validated with intact 3D histology by means of LSFM using a tissue-clearing technique in normal mouse hearts (75) showing a good agreement between both techniques. On the other hand, Teh and colleagues proposed the use of synchrotron radiation imaging (which will be discussed in the next section) to validate DTI measurements of cardiac architecture in fixed healthy rat hearts showing also an excellent agreement in helical and intrusion angles between DTI and structure tensor synchrotron radiation imaging (103).
Finally, some authors have assessed myocardial architecture in T2*-contrast images (77,78) and 3D FLASH images (70,98) using a structure tensor method. Although measurement of helical and intrusion angles are shown to be similar between DTI and T2* and 3D FLASH, the authors reported some differences especially in the ‘sheet’ and ‘sheet-normal’ orientations. More specifically, DTI showed less accuracy in measuring ‘sheet’ orientation as a consequence of poor ‘sheet’ eigenvector assignment. Apart from the lower spatial resolution of DTI, this could be also explained by the fact that in the ventricular myocardium of ex vivo perfused hearts, there may be more than one diffusion component, fast and slow, whereas a single diffusion compartment model in each image voxel is normally used in DTI. Therefore, the authors argue that the proposed orthotropic diffusion may be complicated by non-orthotropic fast diffusion which could result in inaccurate measures of ‘sheet’ orientation (70).
X-ray micro-computed tomography imaging
X-ray imaging, based on material absorption coefficient, is used in the clinics for several cardiovascular applications. Generally, chest X-rays and CT are used to observe heart size and morphology, while coronary angiography with the aid of contrast agents allows for detection of coronary disease. However, the lower contrast of soft-tissue with X-rays compared to MRI and the use of ionizing radiation, relegates it to a non-first-choice technique in most applications (104,105). Recently, the phase shift of X-rays passing through a specimen has been used as an imaging contrast methodology, which is known as X-ray phase contrast imaging (X-PCI). X-PCI is based on the difference in refractive index between different materials, which for soft tissues is actually much larger than in the case of absorption coefficients.
X-rays offer the possibility to increase image resolution in 3D to the micrometer-level (hence the name micro-CT), while keeping short acquisition times, thanks to the use of more brilliant X-ray sources, thus becoming a powerful imaging technique for the detailed analysis of myocardial architecture. In addition, the exploitation of the refractive (phase contrast) and diffractive (scattering) properties of X-ray beams allows to assess cardiac tissue structures, which could otherwise not be resolved with conventional absorption contrast.
In terms of X-ray sources, we can distinguish between (laboratory) X-ray tubes and synchrotron sources, which are mainly differentiated by availability, level of brilliance, acquisition techniques and sample preparation as discussed below.
Micro-computed tomography
Laboratory micro-CT is achieved with compact laboratory X-ray sources. The optimization of the X-ray source and detectors used, as well as the smaller bore, allows to increase resolution (down to a few micrometers) while keeping reasonable acquisitions times (<1 h). For biomedical applications, given that X-ray absorption is measured, it often still relies on the use of contrast agents (e.g., iodine-based) to compensate for the low soft tissue contrast (106). More recently, the use of more powerful nano-focused X-ray sources, such as that found in nano-computed tomography (nano-CT) systems have enabled sub-micron spatial resolution imaging thus resolving microstructural tissue components such as microvasculature or individual cells (107,108).
In terms of cardiovascular applications, Badea et al. (109-111) studied LV morphology and volume in mice in vivo but could not quantify myocardial architecture due to the limited resolution achieved (100 µm). The major breakthrough arrived with the visualization and delineation of the cardiac conduction system in 3D, hitherto only visualized with conventional histology, and the quantification of myocytes orientation in isolated, ex vivo, rat and rabbit hearts (112,113). This was possible by an increase in image resolution down to 30 µm and the use of iodine-based contrast agents, which allows better differentiation of cardiac conduction from myocardial tissue.
Micro-CT has also been used to identify structural anomalies in ex vivo human fetuses and isolated fetal hearts with congenital heart disease (CHD), demonstrating its capability to reproduce, and even give additional diagnostic detail, when compared to conventional techniques (114). In addition, the delineation of the conduction system and quantification of myocytes in normal (115) and in CHD post-natal hearts (116) has also been achieved, thus providing value insight for understanding the electrophysiological and contractile performance of the CHD heart, which could aid clinical diagnosis and management of CHD patients.
As already mentioned, all these studies used iodine-based contrast agents to improve the absorption contrast within myocardial tissue. Nevertheless, a new generation of X-ray sources (liquid metal jet tubes) allow to achieve higher fluxes and partial coherence, which is key for the exploitation of PCI (discussed below). In the work of Reichardt et al. (117), different quantitative structural parameters describing the 3D myocardial architecture were computed in mouse hearts using propagation-based (PB) X-PCI. Even if image quality and resolution (about 5.5 µm voxel size), were highly improved with this new generation of X-ray sources, this approach still entailed dedicated sample preparation and contrast agent combinations.
Current studies show that micro-CT is capable of imaging of a broad range of sample sizes, from murine (~0.5 cm3) to adult human hearts (~575 cm3). Nevertheless, there is a trade-off between sample size and spatial resolution. Larger samples and higher resolution require an increase in radiation deposition and scan time. Therefore, imaging of ex vivo large samples limits the achievable resolution, and vive versa, due to constraints in radiation damage and acquisition time. For in vivo imaging, such limitations still apply and become even more restricted if the experiment is not designed to be terminal.
Synchrotron X-ray phase contrast imaging
Synchrotron light sources are large-scale facilities in which near-speed-of-light electron bundles generate very brilliant and coherent X-ray beams while crossing a series of magnetic fields. Thanks to the highly coherent and brilliant X-ray beams, absorption and phase contrast micro-CT experiments on soft tissues can be routinely performed down to <1 µm isotropic resolution.
In order to perform SR X-PCI, two different approaches are mainly used: grating interferometry (GI) and PB imaging. GI involves the use of gratings but can provide three different images at once after post-processing: absorption, differential phase contrast and dark-field signal. On the contrary, PB can achieve higher resolution with a simpler and faster setup but relies on phase retrieval algorithms in order to provide phase contrast images.
There are some studies that have used GI for the detailed visualization of the gross cardiac morphology of fetal hearts and, particularly, the delineation of conduction system in fetal and neonatal human hearts with CHD (19,118,119). On the other hand, PB X-PCI has been successfully applied for the ex vivo assessment of myocardial architecture in detail in LV transmural pieces of human hearts (17,18,120,121), in whole hearts of fetuses with CHD (15) as well as in hearts from different animal models (13,14). Moreover, PB X-PCI has also shown its potential for the detailed visualization and quantification of coronary arteries, both in human fetuses and in a rabbit model of intrauterine growth restriction (14), as well as the delineation of the conduction system in a fetal heart with a complex CHD without the use of any contrast agent (15).
All the above-mentioned studies focus on the assessment of different macro-structural tissue components, such as myocyte orientation or coronary arteries, both in healthy and in diseased hearts. Nevertheless, it is well known that remodeling also affects the heart at cell and even subcellular level. Recently, Dejea et al. presented a multi-scale PB X-PCI protocol to study the cardiac architecture non-destructively at two different resolutions and without any sample manipulation between the two scans. This approach was applied to investigate detailed cardiac remodeling in rat models of fibrosis and myocardial infarct (13). Lower resolution scans (5.8 µm pixel size) were used to assess gross anatomy, myocyte orientation and coronary arteries, while zoom-in scans of regions of interest with higher resolution (0.65 µm pixel size) were used to quantify collagen, segment individual myocytes and microvasculature and quantify the orientation of myocytes in more detail at an unprecedent resolution (see Figure 2).
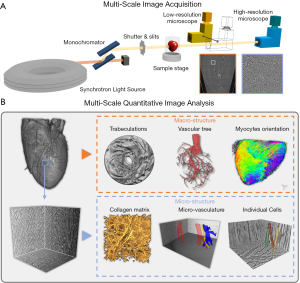
SR X-PCI is thus a very powerful technique for the multi-scale investigation of cardiac architecture thanks to its high-resolution (<1 µm), 3D, non-destructive and time-efficient nature. Nevertheless, this technique is limited to small samples, such as rodent or fetal and neonatal human hearts. As in micro-CT, a trade-off between sample size and spatial resolution, which will determine scan time and dose deposition, must be considered when planning an experiment. Finally, access to synchrotron facilities is limited due to high competition and low availability of experimental slots.
Multimodal imaging
In the previous sections, the capabilities of different imaging techniques to investigate ventricular architecture have been discussed. It was shown that each of these techniques provides different descriptors of cardiac tissue and at different scales, meaning that their combination has the potential to achieve a multi-scale and broader assessment of myocardial architecture. For this reason, multimodal imaging is currently one of the main research topics in the biomedical imaging field.
In the cardiovascular field, multimodal imaging combines techniques at different scales for multi-scale structural visualization and, occasionally, with techniques that can provide quantitative information on myocardial architecture. For example, in the study proposed by Pieles et al., whole mice embryos were first imaged using micro-MRI and, afterwards, individual organs were visualized with HREM (122). Goergen et al. performed an in vivo multi-scale study of myocardial infarcts in mice using (I) molecular MRI to detect apoptosis and necrosis, (II) DT-MRI to compute myofiber orientation, and (III) fluorescence to detect macrophages infiltration in healing myocardium (123). Desgrange et al. presented a multimodal imaging pipeline for phenotyping laterality defects and associated heart malformations in mouse. First, micro-US was used to image the embryonic heart loop in utero, then micro-CT was used for the analysis of organ situs without dissection and finally, HREM was used to assess cardiac left/right anatomy (124). Finally, Rykiel et al. proposed a multi-scale pipeline for the study of remodeling in CHD hearts where whole hearts were imaged with micro-CT and regions of interest were extracted and further imaged with scanning electron microscopy (125).
Discussion and future directions
This paper provides an overview of current and emerging imaging modalities used to investigate myocardial architecture from myocyte to whole organ level. Despite the fact that ventricular architecture has been investigated over several centuries, our understanding of its structure in the healthy heart and changes that occur during a cardiac cycle remain incomplete with ongoing controversy (2,5-7). Myocardial architecture differs between the normal LV and RV. The mature LV has a thick compact layer with myocytes arranged in predominant directions transmurally and fine trabeculations, while the RV is predominantly trabeculated, with only a thin outermost compact layer (7,8). While the myocardial architecture of the LV has been extensively studied, reflected by the large volume of published data, less attention has been paid to the RV. Relatively few articles have characterized the myocardial architecture of the RV (71,94) and quantified changes in different disease states, such as pulmonary hypertension (88) or RV hypertrophy (100), which supports the critical role of RV in determining outcomes in multiple CVDs.
Over the past decades, there have been remarkable improvements in cardiovascular imaging, which have augmented analysis of cardiac structure and function at different scales. Microscopy as well as imaging modalities such as OCT (22) offer high image resolution but need sample preparation, such as fixation and dehydration, that can easily distort tissue and cannot be used to assess tissue microstructure in intact organs or in vivo. US (126) and DT-MRI (68) are the most popular imaging tools for evaluating cardiac structure and function as one. US allows for non-invasive real-time visualization of the heart and blood flow using Doppler imaging, but it does not have the tissue contrast and spatial resolution necessary to investigate cardiac microstructure. MRI offers better contrast than US and differentiates soft tissues better than CT although it requires long acquisition times to get the resolution necessary to assess ventricular architecture. DT-MRI emerged as a promising imaging tool for revealing myocardial architecture in 3D such as myocyte aggregate orientation, fibrosis, etc. However, the long acquisition times needed to achieve high resolution necessary to investigate cardiac architecture in detail, limit its application in vivo. The few in vivo MRI studies achieved only millimeter resolution.
X-ray micro-CT imaging, specially using synchrotron-generated X-rays, has been shown to have histological capabilities in 3D, non-destructively and with relatively short scanning times (<1 h) (127). Recently it has been demonstrated that with synchrotron PB X-PCI is now possible to perform multi-resolution acquisitions on the same sample without any sample manipulation, thus allowing the assessment of overall cardiac geometry and architecture as well as detailed morphological evaluation of structure within focused areas. Nevertheless, most of these studies have been focused on ex vivo non-beating small hearts (13) or had to compromise resolution to achieve in vivo (128) or larger sample sizes. This is mainly due to setup complexity and dose deposition on living animals (128,129). With the advent of ultra-fast imaging (130), the use of more efficient optics (131) and the upgrade into even more powerful/brilliant fourth-generation synchrotrons, scan times can be dramatically reduced and open the door to perform 4D imaging of in vitro and/or in vivo rodent hearts. Due to the limited availability of synchrotron facilities, translation of phase contrast techniques into clinical or laboratory systems is paramount. For example, such translation has already started in other fields such as mammography for breast cancer detection (132), advancing towards in vivo assessment of tissue structure, fibrosis, and vessels without the use of contrast agents. The use of more powerful sources (i.e., liquid jet tubes) and the inclusion of gratings (laboratory GI) will allow the use of X-PCI in a clinical setting. Contrast agents can be avoided, and better in vivo image quality and resolution achieved.
A better understanding of how the microstructural tissue elements of the heart are arranged and integrated together at the whole organ level is crucial to improve our knowledge of cardiac function and dysfunction in health and disease, as well as to develop more realistic and sophisticated computational cardiovascular models that can integrate all imaging information at different scales (133-135). Therefore, we believe that the future of cardiovascular imaging will undoubtedly involve the combination of multiple imaging modalities in order to provide integrative information about the structure and function of the heart and vasculature thus improving the diagnosis of CVDs with the ultimate aim of improving patient care.
Acknowledgments
Funding: PGC has received funding from the postdoctoral fellowships programme Beatriu de Pinos (2018-BP-00201), funded by the Secretary of Universities and Research (Government of Catalonia) and by the Horizon 2020 programme of research and innovation of the European Union under the Marie Skłodowska-Curie grant agreement Nº 801370. HD was supported by the grant #2017-303 of the Strategic Focal Area “Personalized Health and Related Technologies (PHRT)” of the ETH Domain.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Martin Koestenberger, Harm-Jan Bogaard and Georg Hansmann) for the series “Right Ventricular Dysfunction” published in Cardiovascular Diagnosis and Therapy. The article was sent for external peer review organized by the Editor-in-Chief and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cdt-20-269). The series “Right Ventricular Dysfunction” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Walker CA, Spinale FG. The structure and function of the cardiac myocyte: a review of fundamental concepts. J Thorac Cardiovasc Surg 1999;118:375-82. [Crossref] [PubMed]
- Anderson RH, Niederer PF, Sanchez-Quintana D, et al. How are the cardiomyocytes aggregated together within the walls of the left ventricular cone? J Anat 2019;235:697-705. [Crossref] [PubMed]
- LeGrice I, Pope A, Smaill B. The architecture of the heart: myocyte organization and the cardiac extracellular matrix. In: Villarreal FJ. Interstitial fibrosis in heart failure. New York: Springer, 2005:3-21.
- LeGrice IJ, Smaill BH, Chai LZ, et al. Laminar structure of the heart: ventricular myocyte arrangement and connective tissue architecture in the dog. Am J Physiol 1995;269:H571-82. [PubMed]
- MacIver DH, Stephenson RS, Jensen B, et al. The end of the unique myocardial band: Part I. Anatomical considerations. Eur J Cardiothorac Surg 2018;53:112-9. [Crossref] [PubMed]
- Torrent-Guasp F, Ballester M, Buckberg GD, et al. Spatial orientation of the ventricular muscle band: physiologic contribution and surgical implications. J Thorac Cardiovasc Surg 2001;122:389-92. [Crossref] [PubMed]
- Anderson RH, Ho SY, Redmann K, et al. The anatomical arrangement of the myocardial cells making up the ventricular mass. Eur J Cardiothorac Surg 2005;28:517-25. [Crossref] [PubMed]
- Walker LA, Buttrick PM. The right ventricle: biologic insights and response to disease. Curr Cardiol Rev 2009;5:22-8. [Crossref] [PubMed]
- Veeraraghavan R, Poelzing S, Gourdie RG. Intercellular electrical communication in the heart: a new, active role for the intercalated disk. Cell Commun Adhes 2014;21:161-7. [Crossref] [PubMed]
- Rohr S. Role of gap junctions in propagation of the cardiac action potential. Cardiovascular Research 2004;62:309-22. [Crossref] [PubMed]
- Lunkenheimer PP, Redmann K, Kling N, et al. Three-dimensional architecture of the left ventricular myocardium. Anat Rec A Discov Mol Cell Evol Biol 2006;288:565-78. [Crossref] [PubMed]
- Anderson RH, Smerup M, Sanchez-Quintana D, et al. The three-dimensional arrangement of the myocytes in the ventricular walls. Clin Anat 2009;22:64-76. [Crossref] [PubMed]
- Dejea H, Garcia-Canadilla P, Cook AC, et al. Comprehensive analysis of animal models of cardiovascular disease using multiscale X-ray phase contrast tomography. Sci Rep 2019;9:6996. [Crossref] [PubMed]
- Gonzalez-Tendero A, Zhang C, Balicevic V, et al. Whole heart detailed and quantitative anatomy, myofibre structure and vasculature from X-ray phase-contrast synchrotron radiation-based micro computed tomography. Eur Heart J Cardiovasc Imaging 2017;18:732-41. [Crossref] [PubMed]
- Garcia-Canadilla P, Dejea H, Bonnin A, et al. Complex congenital heart disease associated with disordered myocardial architecture in a midtrimester human fetus. Circ Cardiovasc Imaging 2018;11:e007753. [Crossref] [PubMed]
- Dejea H, Garcia-Canadilla P, Stampanoni M, et al. Microstructural analysis of cardiac endomyocardial biopsies with synchrotron radiation-based X-ray phase contrast imaging. In: Pop M, Wright GA. editors. Functional Imaging and Modeling of the Heart. Cham: Springer, 2017:23-31.
- Mirea I, Varray F, Zhu YM, et al. Very high-resolution imaging of post-mortem human cardiac tissue using X-ray phase contrast tomography. In: van Assen H, Bovendeerd P, Delhaas T. editors. Functional Imaging and Modeling of the Heart. Cham: Springer, 2015:172-9.
- Varray F, Mirea I, Langer M, et al. Extraction of the 3D local orientation of myocytes in human cardiac tissue using X-ray phase-contrast micro-tomography and multi-scale analysis. Med Image Anal 2017;38:117-32. [Crossref] [PubMed]
- Kaneko Y, Shinohara G, Hoshino M, et al. Intact imaging of human heart structure Using X-ray phase-contrast tomography. Pediatr Cardiol 2017;38:390-3. [Crossref] [PubMed]
- Lee WN, Pernot M, Couade M, et al. Mapping myocardial fiber orientation using echocardiography-based shear wave imaging. IEEE Trans Med Imaging 2012;31:554-62. [Crossref] [PubMed]
- Papadacci C, Finel V, Provost J, et al. Imaging the dynamics of cardiac fiber orientation in vivo using 3D Ultrasound Backscatter Tensor Imaging. Sci Rep 2017;7:830. [Crossref] [PubMed]
- Huang D, Swanson EA, Lin CP, et al. Optical coherence tomography. Science 1991;254:1178-81. [Crossref] [PubMed]
- Gan Y, Fleming CP. Extracting three-dimensional orientation and tractography of myofibers using optical coherence tomography. Biomed Opt Express 2013;4:2150-65. [Crossref] [PubMed]
- Castonguay A, Lefebvre J, Pouliot P, et al. Serial optical coherence scanning reveals an association between cardiac function and the heart architecture in the aging rodent heart. Biomed Opt Express 2017;8:5027-38. [Crossref] [PubMed]
- Wang Y, Yao G. Optical tractography of the mouse heart using polarization-sensitive optical coherence tomography. Biomed Opt Express 2013;4:2540-5. [Crossref] [PubMed]
- Wang Y, Zhang K, Wasala NB, et al. Histology validation of mapping depth-resolved cardiac fiber orientation in fresh mouse heart using optical polarization tractography. Biomed Opt Express 2014;5:2843-55. [Crossref] [PubMed]
- St Croix CM, Shand SH, Watkins SC. Confocal microscopy: comparisons, applications, and problems. Biotechniques 2005;39:S2-5. [Crossref] [PubMed]
- Sands GB, Gerneke DA, Hooks DA, et al. Automated imaging of extended tissue volumes using confocal microscopy. Microsc Res Tech 2005;67:227-39. [Crossref] [PubMed]
- Tobita K, Garrison JB, Liu LJ, et al. Three-dimensional myofiber architecture of the embryonic left ventricle during normal development and altered mechanical loads. Anat Rec A Discov Mol Cell Evol Biol 2005;283:193-201. [Crossref] [PubMed]
- Ghafaryasl B, Bijnens BH, van Vliet E, et al. Cardiac microstructure estimation from multi-photon confocal microscopy images. In: Ourselin S, Rueckert D, Smith N. editors. Functional Imaging and Modeling of the Heart. Heidelberg: Springer, 2013:80-8.
- Sands GB, Smaill BH, LeGrice IJ. Virtual sectioning of cardiac tissue relative to fiber orientation. Annu Int Conf IEEE Eng Med Biol Soc 2008;2008:226-9. [Crossref] [PubMed]
- Pope AJ, Sands GB, Smaill BH, et al. Three-dimensional transmural organization of perimysial collagen in the heart. Am J Physiol Heart Circ Physiol 2008;295:H1243-52. [Crossref] [PubMed]
- Sands G, Goo S, Gerneke D, et al. The collagenous microstructure of cardiac ventricular trabeculae carneae. J Struct Biol 2011;173:110-6. [Crossref] [PubMed]
- LeGrice IJ, Pope AJ, Sands GB, et al. Progression of myocardial remodeling and mechanical dysfunction in the spontaneously hypertensive rat. Am J Physiol Heart Circ Physiol 2012;303:H1353-65. [Crossref] [PubMed]
- Khwaounjoo P, LeGrice IJ, Trew ML, et al. Quantifying structural and functional differences between normal and fibrotic ventricles. In: van Assen H, Bovendeerd P, Delhaas T. editors. Functional Imaging and Modeling of the Heart. Cham: Springer, 2015:48-56.
- Wang VY, Niestrawska JA, Wilson AJ, et al. Image-driven constitutive modeling of myocardial fibrosis. Int J Comput Methods Eng Sci Mech 2016;17:211-21. [Crossref]
- Weninger WJ, Geyer SH, Mohun TJ, et al. High-resolution episcopic microscopy: A rapid technique for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology. Anat Embryol (Berl) 2006;211:213-21. [Crossref] [PubMed]
- Weninger WJ, Maurer-Gesek B, Reissig LF, et al. Visualising the cardiovascular system of embryos of biomedical model organisms with high resolution episcopic microscopy (HREM). J Cardiovasc Dev Dis 2018. [Crossref] [PubMed]
- Mohun TJ, Weninger WJ. Imaging heart development using high-resolution episcopic microscopy. Curr Opin Genet Dev 2011;21:573-8. [Crossref] [PubMed]
- Geyer SH, Weninger WJ. High-resolution episcopic microscopy (HREM): Looking back on 13 years of successful generation of digital volume data of organic material for 3D visualisation and 3D display. Appl Sci 2019;9:3826. [Crossref]
- Matsui H, Mohun T, Gardiner HM. Three-dimensional reconstruction imaging of the human foetal heart in the first trimester. Eur Heart J 2010;31:415. [Crossref] [PubMed]
- Matsui H, Ho SY, Mohun TJ, et al. Postmortem high-resolution episcopic microscopy (HREM) of small human fetal hearts. Ultrasound Obstet Gynecol 2015;45:492-3. [Crossref] [PubMed]
- Weninger WJ, Geyer SH. Three-dimensional (3D) visualisation of the cardiovascular system of mouse embryos and fetus. Open Cardiovasc Imaging J 2009;1:1-12. [Crossref]
- Anderson RH, Mohun TJ, Brown NA. Clarifying the morphology of the ostium primum defect. J Anat 2015;226:244-57. [Crossref] [PubMed]
- Geyer SH, Reissig LF, Hüsemann M, et al. Morphology, topology and dimensions of the heart and arteries of genetically normal and mutant mouse embryos at stages S21-S23. J Anat 2017;231:600-14. [Crossref] [PubMed]
- Le Garrec JF, Domínguez JN, Desgrange A, et al. A predictive model of asymmetric morphogenesis from 3D reconstructions of mouse heart looping dynamics. Elife 2017. [Crossref] [PubMed]
- Dunlevy L, Bennett M, Slender A, et al. Down’s syndrome-like cardiac developmental defects in embryos of the transchromosomic Tc1 mouse. Cardiovasc Res 2010;88:287-95. [Crossref] [PubMed]
- Lana-Elola E, Watson-Scales S, Slender A, et al. Genetic dissection of Down syndrome- associated congenital heart defects using a new mouse mapping panel. Elife 2016. [Crossref] [PubMed]
- Captur G, Wilson R, Bennett MF, et al. Morphogenesis of myocardial trabeculae in the mouse embryo. J Anat 2016;229:314-25. [Crossref] [PubMed]
- Captur G, Ho CY, Schlossarek S, et al. The embryological basis of subclinical hypertrophic cardiomyopathy. Sci Rep 2016;6:27714. [Crossref] [PubMed]
- Paun B, Bijnens B, Cook AC, et al. Quantification of the detailed cardiac left ventricular trabecular morphogenesis in the mouse embryo. Med Image Anal 2018;49:89-104. [Crossref] [PubMed]
- Garcia-Canadilla P, Cook AC, Mohun TJ, et al. Myoarchitectural disarray of hypertrophic cardiomyopathy begins pre-birth. J Anat 2019;235:962-76. [Crossref] [PubMed]
- Oldenbourg R. Polarized light microscopy: principles and practice. Cold Spring Harb Protoc 2013;2013:1023-36. [Crossref] [PubMed]
- Whittaker P, Boughner DR, Kloner RA. Analysis of healing after myocardial infarction using polarized light microscopy. Am J Pathol 1989;134:879-93. [PubMed]
- de Souza RR. Aging of myocardial collagen. Biogerontology 2002;3:325-35. [Crossref] [PubMed]
- Mendes ABL, Ferro M, Rodrigues B, et al. Quantification of left ventricular myocardial collagen system in children, young adults, and the elderly. Medicina (B Aires) 2012;72:216-20. [PubMed]
- Ohayon J, Usson Y, Jouk PS, et al. Fibre orientation in human fetal heart and ventricular mechanics: a small perturbation. Comput Methods Biomech Biomed Engin 1999;2:83-105. [Crossref] [PubMed]
- Jouk PS, Usson Y, Michalowicz G, et al. Three-dimensional cartography of the pattern of the myofibres in the second trimester fetal human heart. Anat Embryol (Berl) 2000;202:103-18. [Crossref] [PubMed]
- Jouk PS, Mourad A, Milisic V, et al. Analysis of the fiber architecture of the heart by quantitative polarized light microscopy. Accuracy, limitations and contribution to the study of the fiber architecture of the ventricles during fetal and neonatal life. Eur J Cardiothorac Surg 2007;31:915-21. [Crossref] [PubMed]
- Santi PA. Light sheet fluorescence microscopy: a review. J Histochem Cytochem 2011;59:129-38. [Crossref] [PubMed]
- Weber M, Huisken J. Light sheet microscopy for real-time developmental biology. Curr Opin Genet Dev 2011;21:566-72. [Crossref] [PubMed]
- Ding Y, Ma J, Langenbacher AD, et al. Multiscale light-sheet for rapid imaging of cardiopulmonary system. JCI Insight 2018;3:e121396. [Crossref] [PubMed]
- Fei P, Nie J, Lee J, et al. Subvoxel light-sheet microscopy for high-resolution high-throughput volumetric imaging of large biomedical specimens. Adv Photonics 2019;1:016002. [Crossref]
- Ding Y, Bailey Z, Messerschmidt V, et al. Light-sheet fluorescence microscopy for the study of the murine heart. J Vis Exp 2018.57769. [Crossref] [PubMed]
- Mickoleit M, Schmid B, Weber M, et al. High-resolution reconstruction of the beating zebrafish heart. Nat Methods 2014;11:919-22. [Crossref] [PubMed]
- Fei P, Lee J, Packard RRS, et al. Cardiac light-sheet fluorescent microscopy for multi-scale and rapid imaging of architecture and function. Sci Rep 2016;6:22489. [Crossref] [PubMed]
- Baek KI, Ding Y, Chang CC, et al. Advanced microscopy to elucidate cardiovascular injury and regeneration: 4D light-sheet imaging. Prog Biophys Mol Biol. 2018;138:105-15. [Crossref] [PubMed]
- Mekkaoui C, Reese TG, Jackowski MP, et al. Diffusion MRI in the heart. NMR Biomed 2017;30:e3426. [Crossref] [PubMed]
- Sosnovik DE, Wang R, Dai G, et al. Diffusion MR tractography of the heart. J Cardiovasc Magn Reson 2009;11:47. [Crossref] [PubMed]
- Bernus O, Radjenovic A, Trew ML, et al. Comparison of diffusion tensor imaging by cardiovascular magnetic resonance and gadolinium enhanced 3D image intensity approaches to investigation of structural anisotropy in explanted rat hearts. J Cardiovasc Magn Reson 2015;17:31. [Crossref] [PubMed]
- Teh I, Burton RA, McClymont D, et al. Mapping cardiac microstructure of rabbit heart in different mechanical states by high resolution diffusion tensor imaging: a proof-of-principle study. Prog Biophys Mol Biol 2016;121:85-96. [Crossref] [PubMed]
- Hoffman MP, Taylor EN, Aninwene GE, et al. Assessing the multiscale architecture of muscular tissue with Q-space magnetic resonance imaging Microsc Res Tech 2018;81:162-70. Review. [Crossref] [PubMed]
- Jiang Y, Pandya K, Smithies O, et al. Three-dimensional diffusion tensor microscopy of fixed mouse hearts. Magn Reson Med 2004;52:453-60. [Crossref] [PubMed]
- Angeli S, Befera N, Peyrat JM, et al. A high-resolution cardiovascular magnetic resonance diffusion tensor map from ex-vivo C57BL/6 murine hearts. J Cardiovasc Magn Reson 2014;16:77. [Crossref] [PubMed]
- Lee SE, Nguyen C, Yoon J, et al. Three-dimensional cardiomyocytes structure revealed by diffusion tensor imaging and its validation using a tissue-clearing technique. Sci Rep 2018;8:6640. [Crossref] [PubMed]
- Hsu EW, Buckley DL, Bui JD, et al. Two-component diffusion tensor MRI of isolated perfused hearts. Magn Reson Med 2001;45:1039-45. [Crossref] [PubMed]
- Köhler S, Hiller KH, Waller C, et al. Investigation of the microstructure of the isolated rat heart: a comparison between T2*- and diffusion-weighted MRI. Magn Reson Med 2003;50:1144-50. [Crossref] [PubMed]
- Köhler S, Hiller KH, Waller C, et al. Visualization of myocardial microstructure using high-resolution T*2 imaging at high magnetic field. Magn Reson Med 2003;49:371-5. [Crossref] [PubMed]
- Chen J, Song SK, Liu W, et al. Remodeling of cardiac fiber structure after infarction in rats quantified with diffusion tensor MRI. Am J Physiol Heart Circ Physiol 2003;285:H946-54. [Crossref] [PubMed]
- Chen J, Liu W, Zhang H, et al. Regional ventricular wall thickening reflects changes in cardiac fiber and sheet structure during contraction: quantification with diffusion tensor MRI. Am J Physiol Heart Circ Physiol 2005;289:H1898-907. [Crossref] [PubMed]
- Hales PW, Burton RAB, Bollensdorff C, et al. Progressive changes in T1, T2 and left-ventricular histo- architecture in the fixed and embedded rat heart. NMR Biomed 2011;24:836-43. [Crossref] [PubMed]
- Gilbert SH, Benoist D, Benson AP, et al. Visualization and quantification of whole rat heart laminar structure using high-spatial resolution contrast-enhanced MRI. Am J Physiol Heart Circ Physiol 2012;302:H287-98. [Crossref] [PubMed]
- Hales PW, Schneider JE, Burton RAB, et al. Histo-anatomical structure of the living isolated rat heart in two contraction states assessed by diffusion tensor MRI. Prog Biophys Mol Biol 2012;110:319-30. [Crossref] [PubMed]
- Teh I, McClymont D, Burton RA, et al. Resolving fine cardiac structures in rats with high-resolution diffusion tensor imaging. Sci Rep 2016;6:30573. [Crossref] [PubMed]
- Scollan DF, Holmes A, Zhang J, et al. Reconstruction of cardiac ventricular geometry and fiber orientation using magnetic resonance imaging. Ann Biomed Eng 2000;28:934-44. [Crossref] [PubMed]
- Tseng WY, Wedeen VJ, Reese TG, et al. Diffusion tensor MRI of myocardial fibers and sheets: correspondence with visible cut-face texture. J Magn Reson Imaging 2003;17:31-42. [Crossref] [PubMed]
- Geerts L, Bovendeerd P, Nicolay K, et al. Characterization of the normal cardiac myofiber field in goat measured with MR-diffusion tensor imaging. Am J Physiol Heart Circ Physiol 2002;283:H139-45. [Crossref] [PubMed]
- Agger P, Lakshminrusimha S, Laustsen C, et al. The myocardial architecture changes in persistent pulmonary hypertension of the newborn in an ovine animal model. Pediatr Res 2016;79:565-74. [Crossref] [PubMed]
- Helm P, Beg MF, Miller M, et al. Measuring and mapping cardiac fiber and laminar architecture using diffusion tensor MR imaging topics review of DTMRI and estimation of cardiac fiber. Ann N Y Acad Sci 2005;1047:296-307. [Crossref] [PubMed]
- Helm PA, Tseng HJ, Younes L, et al. Ex vivo 3D diffusion tensor imaging and quantification of cardiac laminar structure. Magn Reson Med 2005;54:850-9. [Crossref] [PubMed]
- Poveda F, Gil D, Martí E, et al. Helical structure of the cardiac ventricular anatomy assessed by diffusion tensor magnetic resonance imaging with multiresolution tractography. Rev Esp Cardiol (Engl Ed) 2013;66:782-90. [Crossref] [PubMed]
- Schmid P, Jaermann T, Boesiger P, et al. Ventricular myocardial architecture as visualised in postmortem swine hearts using magnetic resonance diffusion tensor imaging. Eur J Cardiothorac Surg 2005;27:468-72. [Crossref] [PubMed]
- Smerup M, Agger P, Nielsen EA, et al. Regional and epi- to endocardial differences in transmural angles of left ventricular cardiomyocytes measured in ex vivo pig hearts: functional implications. Anat Rec (Hoboken) 2013;296:1724-34. [Crossref] [PubMed]
- Omann C, Agger P, Bøgh N, et al. Resolving the natural myocardial remodelling brought upon by cardiac contraction; a porcine ex-vivo cardiovascular magnetic resonance study of the left and right ventricle. J Cardiovasc Magn Reson 2019;21:35. [Crossref] [PubMed]
- Nielles-Vallespin S, Khalique Z, Ferreira PF, et al. Assessment of myocardial microstructural dynamics by in vivo diffusion tensor cardiac magnetic resonance. J Am Coll Cardiol 2017;69:661-76. [Crossref] [PubMed]
- Lohr D, Terekhov M, Weng AM, et al. Spin echo based cardiac diffusion imaging at 7T: an ex vivo study of the porcine heart at 7T and 3T. PLoS One 2019;14:e0213994. [Crossref] [PubMed]
- Mekkaoui C, Porayette P, Jackowski MP, et al. Diffusion MRI tractography of the developing human fetal heart. PLoS One 2013;8:e72795. [Crossref] [PubMed]
- Haliot K, Magat J, Ozenne V, et al. 3D high resolution imaging of human heart for visualization of the cardiac structure. In: Coudière Y, Ozenne V, Vigmond E, et al. editors. Functional Imaging and Modeling of the Heart. Cham: Springer, 2019:196-207.
- Rohmer D, Sitek A, Gullberg GT. Reconstruction and visualization of fiber and laminar structure in the normal human heart from ex vivo diffusion tensor magnetic resonance imaging (DTMRI) data. Invest Radiol 2007;42:777-89. [Crossref] [PubMed]
- Nielsen E, Smerup M, Agger P, et al. Normal right ventricular three-dimensional architecture, as assessed with diffusion tensor magnetic resonance imaging, is preserved during experimentally induced right ventricular hypertrophy. Anat Rec (Hoboken) 2009;292:640-51. [Crossref] [PubMed]
- Scott AD, Ferreira PF, Nielles-Vallespin S, et al. Optimal diffusion weighting for in vivo cardiac diffusion tensor imaging. Magn Reson Med 2015;74:420-30. [Crossref] [PubMed]
- Ferreira PF, Kilner PJ, Mcgill LA, et al. In vivo cardiovascular magnetic resonance diffusion tensor imaging shows evidence of abnormal myocardial laminar orientations and mobility in hypertrophic cardiomyopathy. J Cardiovasc Magn Reson 2014;16:87. [Crossref] [PubMed]
- Teh I, McClymont D, Zdora M-C, et al. Validation of diffusion tensor MRI measurements of cardiac microstructure with structure tensor synchrotron radiation imaging. J Cardiovasc Magn Reson 2017;19:31. [Crossref] [PubMed]
- Stokes MB, Roberts-Thomson R. The role of cardiac imaging in clinical practice. Aust Prescr 2017;40:151-5. [Crossref] [PubMed]
- Watson SR, Dormer JD, Fei B. Imaging technologies for cardiac fiber and heart failure: a review. Heart Fail Rev 2018;23:273-89. [Crossref] [PubMed]
- Metscher BD. MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Dev Dyn 2009;238:632-40. [Crossref] [PubMed]
- Khoury BM, Bigelow EM, Smith LM, et al. The use of nano-computed tomography to enhance musculoskeletal research. Connect Tissue Res 2015;56:106-19. [Crossref] [PubMed]
- Walton LA, Bradley RS, Withers PJ, et al. Morphological characterisation of unstained and intact tissue micro-architecture by X-ray computed micro- and nano-tomography. Sci Rep 2015;5:10074. [Crossref] [PubMed]
- Badea CT, Fubara B, Hedlund LW, et al. 4-D micro-CT of the mouse heart. Mol Imaging 2005;4:110-6. [Crossref] [PubMed]
- Badea CT, Wetzel AW, Mistry N, et al. Left ventricle volume measurements in cardiac micro-CT: The impact of radiation dose and contrast agent. Comput Med Imaging Graph 2008;32:239-50. [Crossref] [PubMed]
- Badea CT, Schreibmann E, Fox T. A registration based approach for 4D cardiac micro-CT using combined prospective and retrospective gating. Med Phys 2008;35:1170-9. [Crossref] [PubMed]
- Stephenson RS, Boyett MR, Hart G, et al. Contrast enhanced micro-computed tomography resolves the 3-dimensional morphology of the cardiac conduction system in mammalian hearts. PLoS One 2012;7:e35299. [Crossref] [PubMed]
- Stephenson RS, Agger P, Lunkenheimer PP, et al. The functional architecture of skeletal compared to cardiac musculature: myocyte orientation, lamellar unit morphology, and the helical ventricular myocardial band. Clin Anat 2016;29:316-32. [Crossref] [PubMed]
- Lombardi CM, Zambelli V, Botta G, et al. Postmortem microcomputed tomography (micro-CT) of small fetuses and hearts. Ultrasound Obstet Gynecol 2014;44:600-9. [Crossref] [PubMed]
- Stephenson RS, Atkinson A, Kottas P, et al. High resolution 3-dimensional imaging of the human cardiac conduction system from microanatomy to mathematical modeling. Sci Rep 2017;7:7188. [Crossref] [PubMed]
- Stephenson RS, Jones CB, Guerrero R, et al. High-resolution contrast-enhanced micro-computed tomography to identify the cardiac conduction system in congenitally malformed hearts: valuable insight from a hospital archive. JACC Cardiovasc Imaging 2018;11:1706-12. [Crossref] [PubMed]
- Reichardt M, Töpperwien M, Khan A, et al. Fiber orientation in a whole mouse heart reconstructed by laboratory phase-contrast micro-CT. J Med Imaging (Bellingham) 2020;7:023501. [Crossref] [PubMed]
- Tsukube T, Yagi N, Hoshino M, et al. Impact of synchrotron radiation-based X-ray phase-contrast tomography on understanding various cardiovascular surgical pathologies. Gen Thorac Cardiovasc Surg 2015;63:590-2. [Crossref] [PubMed]
- Shinohara G, Morita K, Hoshino M, et al. Three dimensional visualization of human cardiac conduction tissue in whole heart specimens by high-resolution phase-contrast CT imaging using synchrotron radiation. World J Pediatr Congenit Heart Surg 2016;7:700-5. [Crossref] [PubMed]
- Mirea I, Wang L, Varray F, et al. Statistical analysis of transmural laminar microarchitecture of the human left ventricle. In: 2016 IEEE 13th International Conference on Signal Processing (ICSP). Chengdu, 2017:53-6.
- Wang S, Mirea I, Varray F, et al. Investigating the 3D local myocytes arrangement in the human LV mid-wall with the transverse angle. In: Coudière Y, Ozenne V, Vigmond E, et al. editors. Functional Imaging and Modeling of the Heart. Cham: Springer, 2019:208-16.
- Pieles G, Geyer SH, Szumska D, et al. microMRI-HREM pipeline for high-throughput, high-resolution phenotyping of murine embryos. J Anat 2007;211:132-7. [Crossref] [PubMed]
- Goergen CJ, Sosnovik DE. From molecules to myofibers: multiscale imaging of the myocardium. J Cardiovasc Transl Res 2011;4:493-503. [Crossref] [PubMed]
- Desgrange A, Lokmer J, Marchiol C, et al. Standardised imaging pipeline for phenotyping mouse laterality defects and associated heart malformations, at multiple scales and multiple stages. Dis Model Mech 2019;12:dmm038356. [Crossref] [PubMed]
- Rykiel G, López CS, Riesterer JL, et al. Multiscale cardiac imaging: from whole heart images to cardiac ultrastructure. Microsc Microanal 2019;25:1198-9. [Crossref]
- Cikes M, Tong L, Sutherland GR, et al. Ultrafast cardiac ultrasound imaging: technical principles, applications, and clinical benefits. JACC Cardiovasc Imaging 2014;7:812-23. [Crossref] [PubMed]
- Zehbe R, Haibel A, Riesemeier H, et al. Going beyond histology. Synchrotron micro-computed tomography as a methodology for biological tissue characterization: from tissue morphology to individual cells. J R Soc Interface 2010;7:49-59. [Crossref] [PubMed]
- Lovric G, Barré SF, Schittny JC, et al. Dose optimization approach to fast X-ray microtomography of the lung alveoli. J Appl Crystallogr 2013;46:856-60. [Crossref] [PubMed]
- Immel A, Le Cabec A, Bonazzi M, et al. Effect of X-ray irradiation on ancient DNA in sub-fossil bones - Guidelines for safe X-ray imaging. Sci Rep 2016;6:32969. [Crossref] [PubMed]
- Mokso R, Schlepütz CM, Theidel G, et al. GigaFRoST: The gigabit fast readout system for tomography. J Synchrotron Radiat 2017;24:1250-9. [Crossref] [PubMed]
- Bührer M, Stampanoni M, Rochet X, et al. High-numerical-aperture macroscope optics for time-resolved experiments. J Synchrotron Radiat 2019;26:1161-72. [Crossref] [PubMed]
- Castelli E, Tonutti M, Arfelli F, et al. Mammography with synchrotron radiation: first clinical experience with phase-detection technique. Radiology 2011;259:684-94. [Crossref] [PubMed]
- Lamata P, Casero R, Carapella V, et al. Images as drivers of progress in cardiac computational modelling. Prog Biophys Mol Biol 2014;115:198-212. [Crossref] [PubMed]
- Lopez-Perez A, Sebastian R, Ferrero JM. Three-dimensional cardiac computational modelling: methods, features and applications. Biomed Eng Online 2015;14:35. [Crossref] [PubMed]
- Niederer SA, Lumens J, Trayanova NA. Computational models in cardiology. Nat Rev Cardiol 2019;16:100-11. [Crossref] [PubMed]


