microRNA-based diagnostics and therapy in cardiovascular disease—Summing up the facts
In 1993 the research community began to discover the function and importance of small RNAs and their power in controlling gene expression in humans (1) and termed them “microRNAs” (miRNAs) (2). Since then miRNAs have attracted scientists’ attention for their promising role as diagnostic and prognostic biomarkers in various diseases (3-5) and have been described as potential gene specific therapeutic targets in disease modeling (6).
In cardiovascular disease, miRNAs are considered disease-specific biomarkers with defined groups of circulating miRNAs being quantitatively altered in certain disease entities such as myocardial infarction (MI), atherosclerosis, coronary artery disease (CAD), heart failure, atrial fibrillation, hypertrophy and fibrosis (7-10).
In this review article we are discussing the value of cardiovascular miRNAs with a focus on CAD and MI and provide an overview about their use as acute and prognostic biomarkers as well as therapeutic agents for cardiovascular diseases.
miRNA—a wrap-up of what they are
miRNAs are a class of abundant, non-coding RNAs of 19-25 nucleotides length recognized as evolutionarily conserved regulatory RNA molecules (11,12). miRNAs negatively regulate gene expression at the post-transcriptional level by translational repression or degradation of messenger RNAs (mRNAs), thus affecting a variety of cell processes (13-15). At the time of writing 2,588 mature homo sapiens miRNAs are listed in “miRBase” (16). Computational prediction of target mRNAs suggests that more than 60% of all mammalian protein-coding genes are conserved targets of miRNAs (17), while every single miRNA has target sites in hundreds of different genes (18). miRNAs reveal tissue-specific expression (19) but also show concentration-dependent effects in pathologically affected organs and tissues (20).
The discovery of microRNAs as promising new biomarkers in the field of cardiovascular disease and CAD in particular has ignited great expectations. miRNA quantification showed organ- and cell-specific expression patterns of certain miRNAs (19) and in vitro findings suggest groups of miRNAs being specifically up and down regulated in different fields of cardiovascular disease (21), while polymorphisms in the miRNA regulatory pathway—so called miRSNPs—have shown association with different types of disease (22-24). Therefore, miRNAs fulfill several criteria of an ideal biomarker. Stability in the circulation, tissue- and phathology-specific regulation as well as high sensitivity and specificity suggest that miRNAs’ applicability as biomarkers for cardiovascular disease might even exceed protein-based biomarkers (25,26). The field of miRNA research has created potential new means of biomarker-based risk stratification for cardiovascular events and promising data have been collected in a large-scale prospective clinical study (27).
Biomarkers in the field of CAD
The diagnosis of stable CAD with an elevated risk for future MI is based on invasive coronary angiography. Studies with CAD patients have revealed altered levels of certain miRNAs compared with healthy controls (7,21,28) highlighting their potential use as diagnostic biomarkers in CAD.
The gain in sensitivity of newly developed high sensitive cardiac troponin (hs-cTn) assay (29,30) comes along with an increase in false positive measurements caused by non-coronary cardiac pathologies (31). Moreover the early diagnosis of acute myocardial infarction (AMI) is still limited by the delayed release of troponin. A biomarker combining high sensitivity and specificity is needed to earlier detect AMI and to further reduce AMI mortality (32). Furthermore, there is still a lack of possibilities to differentiate troponin-negative unstable angina pectoris (UAP) from non-coronary chest pain (NCCP). In this acute setting of cardiovascular disease numerous studies have shown that miRNAs can serve as biomarkers for MI (33-35).
miRNAs in CAD and MI
By modulating different pathways implicated in the regulation of endothelial cell (EC) functions microRNAs and their key regulatory enzyme Dicer play a major role in the function of ECs (36). Dicer-knock down induced dysregulation of miRNAs that are highly expressed in ECs (let-7, mir-21, mir-126, mir-221, and mir-222) results in impaired EC proliferation (37) going along with decreased levels of inflammatory chemokines and cytokines such as IL-8, IL-1β as well as chemokine ligands 1 and 3 (38). These observations emphasize the involvement of miRNAs in the development of atherosclerosis and CAD.
During the process of plaque development different sets of miRNAs have been found in different stages of plaque progression (39) and miRNA dysregulation plays a crucial role in the destabilization and rupture of atherosclerotic plaques (40).
Vascular miRNAs
In vitro studies revealed several miRNAs to specifically influence angiogenesis and the integrity of the endothelial monolayer (37). In particular, miR-27b and miR-130a were detected to stimulate and miR-221 and miR-222 to inhibit angiogenesis (41,42) linking them to the pathogenesis of atherosclerosis and CAD. In fact, miR-221 and -222 have been described to regulate other miRNAs in an in vitro study, amongst them miR-223 (43). The development of atherosclerotic plaque involves inflammation, which is strongly influenced by cholesterol homeostasis (44). In a mouse model Vickers et al. were able to show that cholesterol biosynthesis as well as uptake and efflux are regulated by miR-223 (45) suggesting miR-223 to be a potential mediator of atherosclerosis. Moreover in the same study the authors describe elevated levels of miR-223 inside thrombocytes of patients with diagnosed atherosclerosis. At the same time angiogenesis associated miRNA-106b, -25, -92a and -21 were described as significantly increased in microparticles from atherosclerotic plaques (46). Especially miR-92a had been proven to be a key regulator of angiogenesis in a mouse model as well as in human umbilical vein derived endothelial cells (47). Interestingly though, different studies found pro-atherosclerotic functions of miR-92a and an overexpression of miR-92a in ECs blocked angiogenesis in vitro and in vivo (47,48).
miR-143 and -145 are the most highly expressed microRNAs in vascular smooth muscle cells (VSMCs), controlling differentiation and function (49). In an in vitro model their expression was found decreased during vascular stress suggesting their function in a modulated phenotype of VSMCs as observed in vascular disease (50). To emphasize their importance in atherosclerotic disease, miR-143 and -145 were identified as essential for neointima formation in response to vascular injury (51).
Beside miR-142 and -145, miR-126 was described as blood flow-sensitive miRNA, which regulates endothelial gene expression and is involved in endothelial dysfunction as well as atherosclerosis triggered by alterations of blood flow conditions (52) as they occur in CAD.
Myocardial miRNAs
miR-1 is the most abundant miRNA specific for cardiac and skeletal muscle and functions as a regulator of differentiation and proliferation during cardiogenesis (53) as well as a regulator of cardiomyocyte growth in the adult heart (54). At the same time miR-1 is considered pro-apoptotic in myocardial ischemia (55). In a rat model miR-1 levels significantly increased in the myocardium 12 hours after coronary artery occlucion (56). In human hearts of patients who had died of MI miR-1 was upregulated in remote myocardium as compared to infarcted tissue or healthy adult hearts (57).
miR-133, which is transcribed from the same chromosomal loci as miR-1, enhances myoblast proliferation and thus is involved in cardiomyocyte proliferations (58). Interestingly, though, while miR-1 supports apoptosis in oxidative stress, miR-133 was found to repress caspase-9 expression and thus has anti-apoptotic effects (55).
miR-21 is upregulated in cardiomyocytes shortly after initiation of ischemia before during cell death its concentration decreases (59). However, increased expression of miR-21 in fibroblasts enhances their proliferation (59) suggesting a role of miR-21 in remodeling.
As shown in a mouse model miR-208 is exclusively expressed in cardiomyocytes and consequently released during cardiomyocyte death in MI (60).
Members of the miR-29 family are downregulated in the region adjacent to MI areas in mice and humans (61). The authors discuss the role of miR-29 as a regulator in cardiac fibrosis and a potential target of tissue fibrosis.
Levels of miR-126 were up regulated in the non-infracted areas after induced MI in rat hearts (62). At the same time a different group reported significantly reduced survival rates in miR-126 knock-out mice after permanent coronary artery occlusion compared with wild-type mice (63). The authors hold defective angiogenesis in miR-126-null mice responsible for this effect. miR-126 is supposed to be mainly involved in the reparative phase after myocardial injury by enhancing angiogenesis (59).
The effect of ischemia-reperfusion on miRNAs in the rat heart was evaluated by Tang et al. (64). The authors found that levels of miR-1, miR-126 and miR-208 were increased while miR-21, miR-133 and miR-195 levels had decreased. Botjancic and colleagues analyzed cardiac tissue samples of 50 patients who had died from MI as well as eight formerly healthy trauma victims (65). The authors found miR-208 upregulated in infarcted tissue whereas miR-1, miR-133a/b were downregulated compared with healthy controls.
miRNAs and their use as circulating biomarkers in body fluids
Intracellular miRNAs have been proven to be regulators and mediators of the metabolism under pathological conditions (66). Intercellular miRNAs provide gene regulation and phenotypic control in physiological conditions (67). Having been stably detected in numerous body fluids including serum and plasma as well as saliva, tears and urine (25,26), the importance of miRNAs as extracellular messengers in cell to cell communication and their potential use as circulating biomarkers for cardiovascular disease has been recognized over the past few years (3,68).
Circulating miRNAs have also been detected in exosomes, microvesicles, and apoptotic bodies—so-called microparticles (69,70) —as well as in association with RNA-binding proteins (71) or in HDL lipoprotein complexes (72). These structural conditions are being held responsible for the extraordinary stability of miRNAs in circulating blood (25,73), which presents a prerequisite for the detection of miRNAs in body fluids. The release of miRNAs into extracellular compartments and thus into body fluids and especially into the blood stream has presented the possibility to non-invasively detect those circulating miRNAs and to use them as disease biomarkers. Numerous studies in the field of biomarker research are based on quantification of circulating miRNAs.
Circulating miRNAs in the diagnosis of acute MI
Subsequently to the identification of miR-208 to be exclusively expressed in the hearts of rats Ji et al. used an isopropanol-induced MI model to show that miR-208 plasma levels significantly increased after MI (74). The authors found a comparable time course with cardiac troponin with respect to the detection limit and concluded that miR-208 might serve as a useful circulating biomarker in MI detection. Verification of these findings came from a different study group that found miR-208 plasma levels elevated in rats after coronary artery occlusion (75). In plasma levels of 407 patients with suspected ACS miR-208b and miR-499-5p were tested to successfully discriminate MI (76). Compared with troponin T, the discriminating power measured by AUC was considerable lower (cTnT =0.95; miR-208b =0.82; miR-499-5p =0.79).
On the basis of these findings Adachi et al. performed a miRNA array analysis in various human tissues to identify heart-specific miRNAs and found miR-499 almost specifically expressed in the heart (77), concluding that miR-499 might serve as an additional promising biomarker of MI. Similar results were reported in 92 elderly non ST segment elevation myocardial infarction (NSTEMI) patients. The authors found miR-499-5p levels more than 80-fold increased compared with 99 control persons as well as 81 patients with acute heart failure without MI (78). The power of miR-499-5p in discriminating NSTEMI from control was comparable to cTnT. More importantly, accuracy to differentiate NSTEMI from acute heart failure was higher for miR-499-5p than for cardiac troponins. This observation suggests that miRNAs might potentially be useful biomarkers helping to reduce cardiac troponins’—and especially hs-cTn—rate of false positive results in non-coronary heart disease.
In serum miR-1 was found to be increased by 200-fold with a peak six hours after induced MI in a rat model while levels correlated with infarction size and serum levels of creatine kinase MB (CK-MB) (79) while in a study with 93 acute MI patients and 66 healthy humans significantly higher miR-1 levels were detected in plasma samples of MI patients compared to controls (80). These findings were confirmed by Wang et al. in 33 acute MI patients compared with 30 healthy people as well as 16 patients with stable CAD and 17 patients with other cardiovascular disease (75). In a similar setting plasma miRNA levels of 444 ACS patients were measured and levels of miR-1, miR-133a and miR-208b were elevated in MI compared with patients with UAP (81). Interestingly levels of miR-1, miR-133a/b and miR-208b were independently associated with high sensitive troponin T levels. Li and colleagues also found miR-1, miR-133a, miR-208b and miR-499 plasma levels elevated in 67 patients with acute MI compared to 32 healthy controls (82). At discharge from hospital the expression of the miRNAs had fallen to their baseline plasma levels. Moreover, ROC curve analyses revealed that those four miRNAs were non-inferior to troponin T with respect to prediction of MI.
These data indicate that several circulating miRNA show deregulated levels during ischemic conditions and MI and seem to be suitable biomarkers for the diagnosis of MI. Table 1 gives an overview of circulating miRNAs dysregulated in MI.
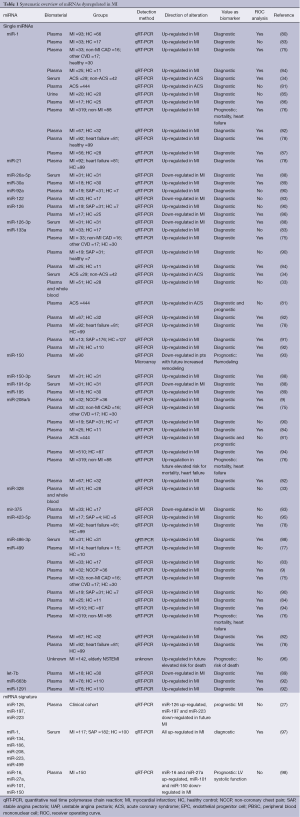
Full table
Recently, in a meta-analysis Cheng et al. concluded that miRNAs are suitable for use as diagnostic biomarkers of MI (99). Fifteen studies with reported sensitivity, specificity and AUC analysis for MI were found while four studies did not report these findings. They found an overall sensitivity of 0.78, specificity of 0.82, and an AUC of 0.91 for overall miRNA analysis with respect to discrimination of MI. Subgroup analysis for the four most-reported miRNAs (miR-1, miR-133a, miR-208b and miR-499) confirmed these findings, whereby miR-1 showed particularly low sensitivity and specificity. Furthermore, the authors found a high correlation between miRNAs and other diagnostic biomarkers of MI such as cTnT, hs-cTnT as well as CK and CK-MB.
Compared with blood samples reports of miRNA quantification for diagnostic purposes of MI in other body fluids are rare. In 20 STEMI patients urine levels of miR-1 were found significantly elevated compared with 20 healthy controls, though (85). Worth noticing is the fact that urine miR-1 levels were reported strongly correlated to serum cardiac troponin I and CK-MB.
Circulating miRNAs in the diagnosis of stable CAD
One of the first groups to study human circulating miRNAs in CAD in a clinical setting was Hoeckstra et al. in 2010 (100). In 50 patients with invasively diagnosed CAD, 129 out of 157 screened miRNAs were expressed in peripheral blood mononuclear cells (PBMCs). miR-135a and -147 were found to be significantly elevated in CAD patients compared with healthy controls. Their findings pointed to the application of blood-based circulating miRNAs as biomarkers in the detection of CAD. Another early case control study has performed a gene expression profiling in whole blood of 163 patients with CAD compared with 63 healthy controls and found miR-140-3p, -182 levels reduced in CAD patients (101).
Fichtlscherer et al. performed a miRNA screening and found the plasma and serum levels of a set of miRNAs altered in 36 patients with documented CAD compared with seventeen healthy controls (7). Interestingly, significantly reduced miRNAs were all either endothelial expressed (miR-126; miR-92a, miR-17) or smooth muscle (miR-145) and inflammatory cell-derived (miR-155). At the same time cardiac muscle enriched circulating miR-133 and miR-208 were elevated in CAD patients. MiR-126, miR-92a and miR-145 had previously been described as promising indicators for atherosclerosis in mouse and in vitro models (see above).
Weber et al. analyzed miRNA levels from whole blood samples of 10 CAD patients compared with 15 healthy controls (102). They found no quantitative changes of endothelial-derived miR-126 and miR-92a as reported before by Fichtlscherer et al. but did detect changes in miR-145 and miR-155—analogous to Fichtlscherer et al. The authors related the discrepancies to underlying differences of the blood samples. While Weber et al. used whole blood, Fichtlscherer et al. analyzed plasma and serum samples. These observations point towards large differences in miRNAs detected in different blood-derived material; these differences become important when considering miRNAs as potential biomarkers for diagnosis.
The finding of dysregulated miR-155 levels in CAD patients were confirmed by observations in 65 CAD patients compared with 22 healthy controls that revealed miR-155 as well as miR-146a significantly induced in CAD patients (103). Elevated levels of miR-146a again were also reported in a recent Chinese case-control study involving 295 CAD patients and 283 healthy controls (104). Independently, significantly higher levels of the same pre-miR-146a variant were associated with CAD in South African patients (105). Finally, in a group of 40 CAD patients compared with healthy controls, platelet rich plasma was analyzed with regard to thrombocyte-derived miRNAs (106). Interestingly, none of the previously described and in atherosclerosis dysregulated platelet-associated miRNAs were described as quantitatively altered between the two groups. Instead miR-340* and miR-634* were upregulated in patients with CAD in two independent validation cohorts. These findings of dysregulated circulating human miRNAs indicate their promising utilization as biomarkers for diagnostic purposes in CAD. More detailed information is given in Table 2.
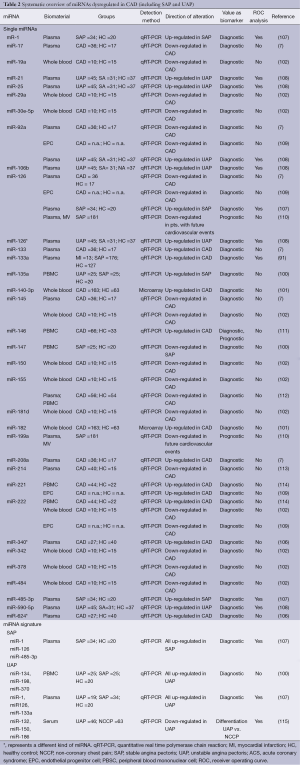
Full table
Circulating miRNAs in the diagnosis of stable and UAP
The definition of UAP includes normal blood values of cardiac troponin. Hence, the diagnosis of UAP is based on clinical evaluation while laboratory parameters are still lacking. Nevertheless, the diagnosis of UAP is crucial since the risk for MI in these patients is elevated. Hoekstra et al. compared miRNA levels derived from PBMCs of 25 UAP patients with those of 25 patients diagnosed with stable angina pectoris (SAP) (100). They found a cluster of three miRNAs (miR-134, miR-198 and miR-370) that was overexpressed in UAP as compared with SAP suggesting that signatures of circulating miRNAs might be useful biomarkers to identify patients at risk for MI. These findings were in line with the description of microparticle derived pro-inflammatory miRNAs that were found to have discriminating power between stable CAD and UAP in the plasma of ten patients (116). However, D’Alessandra et al. failed to confirm these findings in plasma of 19 patients with UAP compared with 34 SAP patients (107). Addressing the same issue Zeller et al. performed a miRNA screening in patients with UAP compared with NCCP followed by a replication phase to verify the results. In a third step the authors evaluated miRNA levels in 46 UAP patients compared with 63 NCCP patients (115). A 3-miRNA-panel of miR-132, miR-150 and miR-186 showed a strong discriminating power between UAP and NCCP. Table 2 includes study results concerning miRNAs in SAP and UAP.
These are promising results on the way to close the diagnostic biomarker-based gap of differentiating stable CAD from ACS in troponin negative patients.
Value of miRNAs in prediction and prognosis of CAD and MI
Beside their diagnostic value circulating miRNAs are also evaluated as potential predictive biomarkers with respect to cardiovascular disease and CAD and MI in particular.
Plasma levels of miR-133a and miR-208b were significantly associated with the risk of death in univariate and age- and gender-adjusted analyses of 444 ACS patients (81). Hoekstra et al. detected a miRNA signature consisting of three miRNAs (miR-134, -198 and -370) with the power to discriminate unstable from stable angina pectoris suggesting a potential to identify patients at risk for future cardiovascular events (100) pointing out the possibilities of miRNAs as a prognostic tool in clinical disease outcome. The finding of miR-370 as a potential prognostic biomarker in risk stratification for acute coronary events is backed up by analogous results in an earlier study in mice. Increased miR-370 expression levels had been found in response to induced ischemia (117). In a study involving 424 patients with suspected MI elevated plasma levels of miR-208b and miR-499-5p were strongly associated with increased risk of mortality or heart failure within 30 days (76). Although the association was lost after adjustment for troponin T the study provides evidence of an association of altered levels of circulating miRNAs with outcome after MI. Recently, in a screening and validation approach miR-652 was found to be significantly associated with post-MI readmission for heart failure while in combination with NT-proBNP and left ventricular ejection fraction this miRNA even improved risk stratification after MI (118). Similar results concerning the risk stratification of mortality or heart failure within 6 months after MI were described for miR-328 and miR-134 in a study population of 359 acute MI patients compared with 30 healthy volunteers (119).
So far, the only known prospective population-based cohort study examining the predictive value of circulating miRNAs with respect to MI included 820 people from the Bruneck study (27). In multivariable Cox regression analysis the authors found a signature of three miRNAs (miR-126, miR-197 and miR-223) involved in the prediction of MI. miR-126 levels were positively while miR-197 and miR-223 inversely associated with future MI. These are promising data for the use of circulating miRNAs in the field of population-based risk assessment for CAD.
MiRNA signatures in CAD
In addition to investigations of single miRNAs, several groups examined the potential of miRNA signatures in cardiovascular disease (Tables 1 and 2).
Ren et al. found a cluster of miRNAs consisting of the miR-106b/25 cluster, miR-17/92a cluster, miR-21/590-5p family, miR-451 as well as miR-126* to be dysregulated in a group of 13 patients with UAP and secured CAD compared with 13 NCCP patients (108). Importantly, even after adjustment for risk factors and the use of statins and anti-platelet drugs the levels of these circulating miRNAs were still independently associated with UAP.
The possibility that specific plasmatic miRNA signatures are capable of efficiently indicating the presence of SAP or UAP compared with healthy controls was supported by findings of D’Alessandra et al. In a cohort of 53 CAD patients with either SAP or UAP the authors found two sets of miRNAs with discriminating power against 20 healthy controls (107). Patients with SAP had higher levels of miR-1, miR-126 and miR-485-3p compared with the control group while miRNA levels in UAP patients were higher in case of miR-1, miR-126 and miR-133a when compared with healthy controls.
MiRNA signatures in acute MI
Li and colleagues examined serum samples from 117 acute MI patients, 182 AP patients and 100 healthy controls (97). After an initial screening of miRNA expression a validation phase revealed a signature consisting of six miRNAs (miR-1, miR-134, miR-186, miR-208, miR-223 and miR-499) with an AUC for the diagnosis of acute MI (AUC =0.83) that was higher than the AUC of cardiac troponin T (AUC =0.768) and CK-MB (0.709) when comparing MI and UAP. Compared with the use of each of the single miRNAs the AUC value for the diagnosis of MI based on the miRNA signature was reported to be progressively higher suggesting superiority of miRNA signatures over single miRNAs. Similar results with respect to miRNA signatures come from Meder et al. In whole blood of patients with AMI they identified 121 miRNAs that were dysregulated compared with controls (120). The authors found a signature of 20 miRNAs that predicts AMI with better specificity and sensitivity as well as higher predictive power than each of the single miRNAs. In their study the use of the miRNA signature even allowed for an earlier diagnosis of AMI than cardiac troponin T. The same study group performed a whole-genome miRNA kinetic study in 18 AMI patients and performed serial measurements of miRNA expression levels (121). They were able to find a signature of seven miRNAs (miR-636, miR-7-1*, miR-380*, miR-1254, miR-455-3p, miR-566, and miR-1291) that was dysregulated early during AMI—five of them being part of the 20 miRNA signature discovered in the previous study. Table 1 provides an overview of miRNA signatures in MI.
Although these studies were performed with rather small sample sizes they emphasize the potential diagnostic power of miRNA signatures as biomarkers in AMI diagnostics.
miRNAs in disease treatment
The functional importance of miRNA-regulated networks is often revealed under stress conditions. At the same time miRNA loss of function may result in only slightly changed phenotypes (66). Difficulties in the use of therapeutically altering miRNAs lie in the fact that a single miRNA can affect expression of numerous proteins and genes while at the same time miRNA based regulation can involve a vast number of different miRNAs. miRNAs impose a relatively modest effect on their target, reflecting that individual mRNAs are targeted by multiple miRNAs while the cellular proteome might be able to compensate the absence of a single miRNA (122). Moreover, miRSNPs that can either modify the sequence of the miRNA itself or the miRNA-target site, are associated not only with cancer risk and outcome but also with altered treatment response (24). Nevertheless, promising results of in vitro testing and animal models indicate the possibility to successfully use miRNAs as therapeutic targets in disease.
Treatment options involving miRNAs imply the idea of specifically influencing the levels of miRNAs in certain diseases. This includes suppression of miRNAs as well as raising miRNA levels or substituting them by artificially generated copies—depending on the mechanism of pathophysiology. The latter can be achieved by the miRNA mimic technology (miR-mimic) which is an approach for gene silencing by generating artificial double-stranded miRNA-like RNA fragments (123). These RNA fragments “mimic” endogenous miRNAs and bind specifically to its target mRNA—activating the RNA-induced silencing complex (RISC), down regulating specific mRNAs and thus induce gene suppression (Figure 1). As opposed to endogenous miRNAs miR-mimics function gene-specifically (123). While in cancer research miR-mimics have already largely been studied, however, in cardiovascular disease only few studies have been published so far.
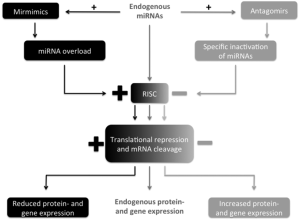
Contrary, miRNA suppression can be obtained by antagomirs (124). Antagomirs are a class of chemically engineered oligonucleotides specifically silencing single endogenous miRNAs (125). They competitively inhibit specific miRNAs by binding to the target mature (124) and lead to a reduced activation of RISC and consequently to an upregulation of specific mRNAs and gene expression (Figure 1) (126).
Further approaches of reducing the amount and effect of miRNAs more or less specifically involve the use of miRNA sponges (also referred to as “target mimicry”), masking and erasers. Sponges are based on the mechanism of blocking the activity of a miRNA by means of a competitive inhibitor that contains a binding site for a miRNA family (127-129). Such inhibitors can be RNAs expressed transiently from transfected plasmids or stably from chromosomal insertions (127). By binding to the miRNA sponge with perfect or imperfect complementarity miRNAs’ effect on mRNA and gene expression is lessened because affected miRNAs cannot bind to their mRNA binding sites (124). As opposed to antagomirs, sponges are specific only to the seed region of a miRNA and thus are capable of blocking a whole family of miRNAs (130).
Masking describes an approach to hinder specific miRNAs from binding to their very binding site by masking the particular mRNA’s binding site of the target miRNA using perfectly matching oligonucleotides (131). As a consequence a smaller amount of miRNAs remains to bind to the target. Erasers are oligonucleotides complementary to a specific miRNA. By binding to the miRNA the eraser inhibits its endogenous function (124).
Studies including miR-mimics
Overexpression of miR-1 induces apoptosis and miR-1 levels strongly correlate with MI size in rats (55,64). By increasing miR-1 levels with the use of ischemic preconditioning a significant reduction of infarction size has been reported (79). miR-1 mimicry has shown that miR-1 regulates endothelin-1 in diabetes (132) but no reports are available specifically for MI.
After miR-21 levels were shown to be reduced in infarcted areas of rats after induced MI, Dong and colleagues were able to inhibit downregulation of this miRNA in infarcted areas by ischemic preconditioning and thus increasing miR-21 levels prior to infarction (62). At the same time transfection with adenovirus expressing miR-21 reduced cell apoptosis, decreased infarction size and improved left ventricular remodeling. Furthermore, intra cardiac injection of miR-21 along with miR-1 and miR-24 in mice was reported to reduce infarct size (133).
A different method of therapeutically raising miRNA levels was described by Huang and coworkers. In an experimental mouse model they showed that transplantation of mesenchymal stem cells overexpressing miR-1 into infarcted myocardium improved cardiac function by enhanced myocyte differentiation compared with control groups (134).
Studies including antagomirs
In a mouse model of MI a systemic application of an antagomir against miR-92a resulted in recovery of damaged tissue by enhanced blood vessel growth (47). Anti-miR-92a antagomirs could induce anti-atherosclerotic effects and block impairment of endothelial cells. An in vitro rat model of induced MI showed that silencing of miR-320 reduced cell death and apoptosis. The inhibition of miR-320 by use of antagomir-320 even led to a significant reduction of infarction size (135).
By using a miR-29 antisense inhibitor in a mouse ischemia-reperfusion model another study group was able to protect cardiomyocytes from injury (136). The authors even succeeded in reducing infarction size by application of antagomirs against miR-29a and miR-29c. miR-92a inhibition by systemic application of anti-miR-92a antagomirs led to enhanced functional recovery of damaged tissue, reduced apoptosis and improved MI size in a mouse model (47). Yang et al. showed that elimination of miR-1 by an antisense inhibitor in infarcted rat hearts had anti-arrhythmogenic effects after induced MI (56).
These studies provide evidence for the use miRNAs as a therapeutic target in CAD and MI and clinical trials are eagerly awaited.
Analytical considerations and standardization
Several confounding factors can influence the quality of miRNA data and need to be critically considered. Different sample preparation approaches and detection and normalization strategies are currently applied in a non-standardized fashion and application of universal reference material is required. Furthermore, variation caused by preanalytical and analytical factors can substantially influence these miRNA data (137). This can particularly be challenging in the clinical setting, as differences in sample collection, sample processing and assay performance in different clinical centers are to be expected. Therefore, to eliminate technical and analytical variability and to avoid artificial data generation, consensus on standard methods for all steps is demanded. Here, we briefly discuss the main influencing factors.
Methodological challenges and influencing factors
RNA isolation
Concentrations of miRNAs in body fluids are low, putting high expectations on the efficiency of their detection. While co-extraction of minimal amounts of inhibiting factors can effect miRNA quantification, it is crucial to find and use methods of miRNA isolation with a maximum of reproducibility and reliability while several studies have described different qualities between the commercially used kits for miRNA isolation (138).
Detection methods
High throughput sequencing, real time PCR (qPCR) and microarrays are the three major quantification methods that are currently being used. While qPCR is considered the “gold standard” in the detection and quantification of defined sets of miRNAs for its high specificity and sensitivity, high-throughput sequencing of miRNAs is superior in the discovery of new miRNAs. Microarrays are considered the best option for standardized genome-wide assays on a larger scale (139). Over the past years universally applicable assays for miRNA quantification have been developed enabling scientists to easily perform qRT-PCR in quantifying miRNA levels. However, for example, in the diagnosis of AMI time is a crucial factor. So far miRNA quantification is still time consuming and bedside testing is not yet possible.
Furthermore, detection of miRNAs compared with the detection of protein-based biomarkers still lacks standardized methods and automatized work flows as well as clear recommendation as to what body fluid to use for what type of miRNA.
Normalization
An important technical aspect concerning miRNA quantification lies in the need for standardized and reproducible isolation and detection protocols as well as uniform means of analyses and normalization in order to efficiently generate robust and reproducible results. Different methods of normalization have been described to have an effect on miRNA quantification (140). There are no standardized control transcripts for normalization, which resulted in the generation of various normalization strategies (141). A widely used method is based on C elegans miR-39 spike in control as a reference miRNA. On the other hand standardization to the mean of all measured miRNAs as well as to individual miRNAs that are detectable at similar levels in all study subjects is highly recommended (142).
Comparing serum and plasma: differing miRNA levels
While some authors reported similar levels of miRNAs in serum and plasma (7,25), inconsistencies between miRNA levels in both materials have also been found (143). Notable is the observation that especially thrombocyte-associated miRNAs in plasma are found in much higher concentrations than in serum (73). Since especially in the field of miRNA-based diagnostics of CAD thrombocyte-derived miRNAs such as miR-126, miR-223 and miR-197 seem to play a significant role in the prognosis of future cardiovascular events (27), standardized methods are needed for future clinical miRNA quantification studies.
Parameters influencing miRNAs levels
Former studies have proven that heparin administered to patients prior to blood sampling interferes with results in miRNA quantification (144,145). Therefore, high caution is needed when selecting patients for in vivo studies of miRNA quantification with respect to heparin administration prior to blood sampling.
Quantification of miRNA levels altered in CAD might also be influenced by the intake of medication such as statins and ACE inhibitors (102).
These findings emphasize the importance to quantify the drug- and metabolite-based influence on miRNAs in the clinical setting. At the same time the inconsistency of the data reflects the necessity of further studies evaluating pathways of how miRNA levels are influenced in circulating blood.
Additionally, it needs to be considered, that levels of biomarkers can also be influenced by the speed of their elimination. Gidlöf et al. found cardiac miRNA levels strongly correlating with renal function indicating that the renal function might also influence the plasma levels of miRNAs (84).
Future steps
Despite a tremendous increase of interest in miRNAs for cardiovascular disease diagnosis and prognosis, we are only just scratching the surface of its complexity. To fully elucidate the potential of miRNAs robust sample processing as well as advances in technology and analysis methods are required.
Discovery of novel miRNAs and confirmation of reported results should be a component of large research networks, involving industry and experts in distinct fields such as molecular biology, analytical chemistry, bioinformatics, clinical-trial design, epidemiology, statistics as well as health-care economics (146). Several collaborative initiatives have emerged in recent years to orchestrate biomarker research efforts. One is the EU funded BiomarCaRE consortium (Biomarker for Cardiovascular Risk Assessment in Europe), a European collaborative research project including over 300,000 participants from 13 European countries in clinical as well as diseased cohorts (147). Amongst other biomarkers miRNAs are evaluated with respect to their predictive value for incident CAD and MI. Quantification of circulating miRNAs on such a large scale has never been performed before and represents the chance to possibly validate findings of the numerous single miRNA studies that have been performed so far.
New players in the field: long non-coding RNAs (lncRNAs)
Besides miRNAs lncRNAs have increasingly attracted attention in the field of gene regulation recently (148). lncRNAs consist of >200 nucleotides and are involved in regulatory mechanisms of gene expression (149). Circulating lncRNAs are being evaluated as biomarkers for cardiovascular disease. First studies revealed differential levels of lncRNAs in blood cells of 414 patients with acute MI (150), suggesting the involvement of lncRNA in pathophysiological conditions of acute MI and their potential future application as circulating cardiovascular biomarkers.
Summary
Numerous studies reported miRNAs as promising new diagnostic and prognostic biomarkers in the field of cardiovascular diseases. The application of circulating miRNAs as biomarkers represents a potential additional module in disease diagnosis and prognosis complimentary to established protein-based biomarkers. In vitro testing and animal models have depicted pathophysiological pathways and authors have developed miRNA-based methods to interfere with disease progression and complications. The use of miR-mimics and antagomirs in the cardiovascular field has not yet found its way into clinical trials but promising study results reflect potential future applications of miRNA therapeutics in this field.
The complex regulatory mechanisms involved in miRNA and gene expression have not yet been completely understood. At the same time technical aspects of detecting and quantifying miRNAs in body fluids including RNA normalization and biomaterial to be used need to be standardized as a basis for reproducible protocols.
Acknowledgements
Funding: T Zeller acknowledges funding by the European Union (BiomarCaRE, grant number: HEALTH-2011-278913), the Deutsche Stiftung für Herzforschung and the Bundesministerium für Bildung und Forschung.
Disclosure: C Schulte reports no conflict of interest.
References
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75:843-54. [PubMed]
- Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science 2001;294:853-8. [PubMed]
- Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit Rev Oncol Hematol 2011;80:193-208. [PubMed]
- Chandrasekaran K, Karolina DS, Sepramaniam S, et al. Role of microRNAs in kidney homeostasis and disease. Kidney Int 2012;81:617-27. [PubMed]
- Fan HM, Sun XY, Guo W, et al. Differential expression of microRNA in peripheral blood mononuclear cells as specific biomarker for major depressive disorder patients. J Psychiatr Res 2014;59:45-52. [PubMed]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 2001;15:188-200. [PubMed]
- Fichtlscherer S, De Rosa S, Fox H, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res 2010;107:677-84. [PubMed]
- Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation 2010;121:1022-32. [PubMed]
- Corsten MF, Dennert R, Jochems S, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet 2010;3:499-506. [PubMed]
- Gomes da Silva AM, Silbiger VN. miRNAs as biomarkers of atrial fibrillation. Biomarkers 2014;19:631-6. [PubMed]
- Lau NC, Lim LP, Weinstein EG, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001;294:858-62. [PubMed]
- Reinhart BJ, Weinstein EG, Rhoades MW, et al. MicroRNAs in plants. Genes Dev 2002;16:1616-26. [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [PubMed]
- Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. Rna 2003;9:277-9. [PubMed]
- Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 2005;310:1817-21. [PubMed]
- Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68-73. [PubMed]
- Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92-105. [PubMed]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol 2007;23:175-205. [PubMed]
- van Rooij E. The art of microRNA research. Circ Res 2011;108:219-34. [PubMed]
- Bauersachs J, Thum T. Biogenesis and regulation of cardiovascular microRNAs. Circ Res 2011;109:334-47. [PubMed]
- Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res 2012;110:483-95. [PubMed]
- Dzikiewicz-Krawczyk A. MicroRNA polymorphisms as markers of risk, prognosis and treatment response in hematological malignancies. Crit Rev Oncol Hematol 2015;93:1-17. [PubMed]
- Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics 2009;10:399-416. [PubMed]
- Salzman DW, Weidhaas JB. SNPing cancer in the bud: microRNA and microRNA-target site polymorphisms as diagnostic and prognostic biomarkers in cancer. Pharmacol Ther 2013;137:55-63. [PubMed]
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8. [PubMed]
- Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010;56:1733-41. [PubMed]
- Zampetaki A, Willeit P, Tilling L, et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol 2012;60:290-9. [PubMed]
- Contu R, Latronico MV, Condorelli G. Circulating microRNAs as potential biomarkers of coronary artery disease: a promise to be fulfilled? Circ Res 2010;107:573-4. [PubMed]
- Wu AH, Jaffe AS. The clinical need for high-sensitivity cardiac troponin assays for acute coronary syndromes and the role for serial testing. Am Heart J 2008;155:208-14. [PubMed]
- Keller T, Zeller T, Peetz D, et al. Sensitive troponin I assay in early diagnosis of acute myocardial infarction. N Engl J Med 2009;361:868-77. [PubMed]
- Hamm CW, Bassand JP, Agewall S, et al. ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation: The Task Force for the management of acute coronary syndromes (ACS) in patients presenting without persistent ST-segment elevation of the European Society of Cardiology (ESC). Eur Heart J 2011;32:2999-3054. [PubMed]
- Li C, Pei F, Zhu X, et al. Circulating microRNAs as novel and sensitive biomarkers of acute myocardial Infarction. Clin Biochem 2012;45:727-32. [PubMed]
- Wang R, Li N, Zhang Y, et al. Circulating microRNAs are promising novel biomarkers of acute myocardial infarction. Intern Med 2011;50:1789-95. [PubMed]
- Kuwabara Y, Ono K, Horie T, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet 2011;4:446-54. [PubMed]
- Jaguszewski M, Osipova J, Ghadri JR, et al. A signature of circulating microRNAs differentiates takotsubo cardiomyopathy from acute myocardial infarction. Eur Heart J 2014;35:999-1006. [PubMed]
- Suárez Y, Fernández-Hernando C, Pober JS, et al. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 2007;100:1164-73. [PubMed]
- Kuehbacher A, Urbich C, Zeiher AM, et al. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res 2007;101:59-68. [PubMed]
- Schroen B, Heymans S. Small but smart--microRNAs in the centre of inflammatory processes during cardiovascular diseases, the metabolic syndrome, and ageing. Cardiovasc Res 2012;93:605-13. [PubMed]
- Jovanović I, Zivković M, Jovanović J, et al. The co-inertia approach in identification of specific microRNA in early and advanced atherosclerosis plaque. Med Hypotheses 2014;83:11-5. [PubMed]
- Menghini R, Stohr R, Federici M. MicroRNAs in vascular aging and atherosclerosis. Ageing Res Rev 2014;17:68-78. [PubMed]
- Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res 2008;79:581-8. [PubMed]
- Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood 2008;111:1217-26. [PubMed]
- Tuccoli A, Poliseno L, Rainaldi G. miRNAs regulate miRNAs: coordinated transcriptional and post-transcriptional regulation. Cell Cycle 2006;5:2473-6. [PubMed]
- Linton MF, Fazio S. Macrophages, inflammation, and atherosclerosis. Int J Obes Relat Metab Disord 2003;27 Suppl 3:S35-40. [PubMed]
- Vickers KC, Landstreet SR, Levin MG, et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci U S A 2014;111:14518-23. [PubMed]
- Zhang J, Ren J, Chen H, et al. Inflammation induced-endothelial cells release angiogenesis associated-microRNAs into circulation by microparticles. Chin Med J (Engl) 2014;127:2212-7. [PubMed]
- Bonauer A, Carmona G, Iwasaki M, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 2009;324:1710-3. [PubMed]
- Wu W, Xiao H, Laguna-Fernandez A, et al. Flow-Dependent Regulation of Kruppel-Like Factor 2 Is Mediated by MicroRNA-92a. Circulation 2011;124:633-41. [PubMed]
- Dahan D, Ekman M, Larsson-Callerfelt AK, et al. Induction of angiotensin converting enzyme after miR-143/145 deletion is critical for impaired smooth muscle contractility. Am J Physiol Cell Physiol 2014;307:C1093-101. [PubMed]
- Elia L, Quintavalle M, Zhang J, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ 2009;16:1590-8. [PubMed]
- Xin M, Small EM, Sutherland LB, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev 2009;23:2166-78. [PubMed]
- Kumar S, Kim CW, Simmons RD, et al. Role of flow-sensitive microRNAs in endothelial dysfunction and atherosclerosis: mechanosensitive athero-miRs. Arterioscler Thromb Vasc Biol 2014;34:2206-16. [PubMed]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005;436:214-20. [PubMed]
- Ikeda S, He A, Kong SW, et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol 2009;29:2193-204. [PubMed]
- Xu C, Lu Y, Pan Z, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci 2007;120:3045-52. [PubMed]
- Yang B, Lin H, Xiao J, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med 2007;13:486-91. [PubMed]
- Bostjancic E, Zidar N, Stajner D, et al. MicroRNA miR-1 is up-regulated in remote myocardium in patients with myocardial infarction. Folia Biol (Praha) 2010;56:27-31. [PubMed]
- Chen JF, Mandel EM, Thomson JM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006;38:228-33. [PubMed]
- Ye Y, Perez-Polo JR, Qian J, et al. The role of microRNA in modulating myocardial ischemia-reperfusion injury. Physiol Genomics 2011;43:534-42. [PubMed]
- van Rooij E, Sutherland LB, Qi X, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 2007;316:575-9. [PubMed]
- van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A 2008;105:13027-32. [PubMed]
- Dong S, Cheng Y, Yang J, et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem 2009;284:29514-25. [PubMed]
- Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 2008;15:261-71. [PubMed]
- Tang Y, Zheng J, Sun Y, et al. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int Heart J 2009;50:377-87. [PubMed]
- Bostjancic E, Zidar N, Stajer D, et al. MicroRNAs miR-1, miR-133a, miR-133b and miR-208 are dysregulated in human myocardial infarction. Cardiology 2010;115:163-9. [PubMed]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012;148:1172-87. [PubMed]
- Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol 2013;33:186-92. [PubMed]
- Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol 2009;21:575-81. [PubMed]
- Zernecke A, Bidzhekov K, Noels H, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal 2009;2:ra81. [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [PubMed]
- Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003-8. [PubMed]
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [PubMed]
- Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997-1006. [PubMed]
- Ji X, Takahashi R, Hiura Y, et al. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem 2009;55:1944-9. [PubMed]
- Wang GK, Zhu JQ, Zhang JT, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 2010;31:659-66. [PubMed]
- Gidlöf O, Smith JG, Miyazu K, et al. Circulating cardio-enriched microRNAs are associated with long-term prognosis following myocardial infarction. BMC Cardiovasc Disord 2013;13:12. [PubMed]
- Adachi T, Nakanishi M, Otsuka Y, et al. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin Chem 2010;56:1183-5. [PubMed]
- Olivieri F, Antonicelli R, Lorenzi M, et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int J Cardiol 2013;167:531-6. [PubMed]
- Cheng Y, Tan N, Yang J, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010;119:87-95. [PubMed]
- Ai J, Zhang R, Li Y, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun 2010;391:73-7. [PubMed]
- Widera C, Gupta SK, Lorenzen JM, et al. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol 2011;51:872-5. [PubMed]
- Li YQ, Zhang MF, Wen HY, et al. Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics (Sao Paulo) 2013;68:75-80. [PubMed]
- D’Alessandra Y, Devanna P, Limana F, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J 2010;31:2765-73. [PubMed]
- Gidlöf O, Andersson P, van der Pals J, et al. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology 2011;118:217-26. [PubMed]
- Duan XX, Zhou X, Wang XB, et al. Urine cardiac specific microRNA-1 level in patients with ST segment elevation acute myocardial infarction. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue 2012;24:709-12. [PubMed]
- Long G, Wang F, Duan Q, et al. Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int J Biol Sci 2012;8:811-8. [PubMed]
- Li LM, Cai WB, Ye Q, et al. Comparison of plasma microRNA-1 and cardiac troponin T in early diagnosis of patients with acute myocardial infarction. World J Emerg Med 2014;5:182-6. [PubMed]
- Hsu A, Chen SJ, Chang YS, et al. Systemic approach to identify serum microRNAs as potential biomarkers for acute myocardial infarction. Biomed Res Int 2014;2014:418628.
- Long G, Wang F, Duan Q, et al. Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS One 2012;7:e50926. [PubMed]
- De Rosa S, Fichtlscherer S, Lehmann R, et al. Transcoronary concentration gradients of circulating microRNAs. Circulation 2011;124:1936-44. [PubMed]
- Wang F, Long G, Zhao C, et al. Plasma microRNA-133a is a new marker for both acute myocardial infarction and underlying coronary artery stenosis. J Transl Med 2013;11:222. [PubMed]
- Peng L, Chun-guang Q, Bei-fang L, et al. Clinical impact of circulating miR-133, miR-1291 and miR-663b in plasma of patients with acute myocardial infarction. Diagn Pathol 2014;9:89. [PubMed]
- Devaux Y, Vausort M, McCann GP, et al. MicroRNA-150: a novel marker of left ventricular remodeling after acute myocardial infarction. Circ Cardiovasc Genet 2013;6:290-8. [PubMed]
- Devaux Y, Vausort M, Goretti E, et al. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin Chem 2012;58:559-67. [PubMed]
- Nabiałek E, Wańha W, Kula D, et al. Circulating microRNAs (miR-423-5p, miR-208a and miR-1) in acute myocardial infarction and stable coronary heart disease. Minerva Cardioangiol 2013;61:627-37. [PubMed]
- Olivieri F, Antonicelli R, Spazzafumo L, et al. Admission levels of circulating miR-499-5p and risk of death in elderly patients after acute non-ST elevation myocardial infarction. Int J Cardiol 2014;172:e276-8. [PubMed]
- Li C, Fang Z, Jiang T, et al. Serum microRNAs profile from genome-wide serves as a fingerprint for diagnosis of acute myocardial infarction and angina pectoris. BMC Med Genomics 2013;6:16. [PubMed]
- Devaux Y, Vausort M, McCann GP, et al. A panel of 4 microRNAs facilitates the prediction of left ventricular contractility after acute myocardial infarction. PLoS One 2013;8:e70644. [PubMed]
- Cheng C, Wang Q, You W, et al. MiRNAs as biomarkers of myocardial infarction: a meta-analysis. PLoS One 2014;9:e88566. [PubMed]
- Hoekstra M, van der Lans CA, Halvorsen B, et al. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem Biophys Res Commun 2010;394:792-7. [PubMed]
- Taurino C, Miller WH, McBride MW, et al. Gene expression profiling in whole blood of patients with coronary artery disease. Clin Sci (Lond) 2010;119:335-43. [PubMed]
- Weber M, Baker MB, Patel RS, et al. MicroRNA Expression Profile in CAD Patients and the Impact of ACEI/ARB. Cardiol Res Pract 2011;2011:532915.
- Zhu J, Chen T, Yang L, et al. Regulation of microRNA-155 in atherosclerotic inflammatory responses by targeting MAP3K10. PLoS One 2012;7:e46551. [PubMed]
- Xiong XD, Cho M, Cai XP, et al. A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutat Res Fundam Mol Mech Mutagen 2014;761:15-20. [PubMed]
- Ramkaran P, Khan S, Phulukdaree A, et al. miR-146a polymorphism influences levels of miR-146a, IRAK-1, and TRAF-6 in young patients with coronary artery disease. Cell Biochem Biophys 2014;68:259-66. [PubMed]
- Sondermeijer BM, Bakker A, Halliani A, et al. Platelets in patients with premature coronary artery disease exhibit upregulation of miRNA340* and miRNA624*. PLoS One 2011;6:e25946. [PubMed]
- D’Alessandra Y, Carena MC, Spazzafumo L, et al. Diagnostic potential of plasmatic MicroRNA signatures in stable and unstable angina. PLoS One 2013;8:e80345. [PubMed]
- Ren J, Zhang J, Xu N, et al. Signature of circulating microRNAs as potential biomarkers in vulnerable coronary artery disease. PLoS One 2013;8:e80738. [PubMed]
- Zhang Q, Kandic I, Kutryk MJ. Dysregulation of angiogenesis-related microRNAs in endothelial progenitor cells from patients with coronary artery disease. Biochem Biophys Res Commun 2011;405:42-6. [PubMed]
- Jansen F, Yang X, Proebsting S, et al. MicroRNA Expression in Circulating Microvesicles Predicts Cardiovascular Events in Patients With Coronary Artery Disease. J Am Heart Assoc 2014;3:e001249. [PubMed]
- Takahashi Y, Satoh M, Minami Y, et al. Expression of miR-146a/b is associated with the Toll-like receptor 4 signal in coronary artery disease: effect of renin-angiotensin system blockade and statins on miRNA-146a/b and Toll-like receptor 4 levels. Clin Sci (Lond) 2010;119:395-405. [PubMed]
- Zhu GF, Yang LX, Guo RW, et al. microRNA-155 is inversely associated with severity of coronary stenotic lesions calculated by the Gensini score. Coron Artery Dis 2014;25:304-10. [PubMed]
- Lu HQ, Liang C, He ZQ, et al. Circulating miR-214 is associated with the severity of coronary artery disease. J Geriatr Cardiol 2013;10:34-8. [PubMed]
- Minami Y, Satoh M, Maesawa C, et al. Effect of atorvastatin on microRNA 221 / 222 expression in endothelial progenitor cells obtained from patients with coronary artery disease. Eur J Clin Invest 2009;39:359-67. [PubMed]
- Zeller T, Keller T, Ojeda F, et al. Assessment of microRNAs in patients with unstable angina pectoris. Eur Heart J 2014;35:2106-14. [PubMed]
- Diehl P, Fricke A, Sander L, et al. Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res 2012;93:633-44. [PubMed]
- Xu CF, Yu CH, Li YM. Regulation of hepatic microRNA expression in response to ischemic preconditioning following ischemia/reperfusion injury in mice. Omics 2009;13:513-20. [PubMed]
- Pilbrow AP, Cordeddu L, Cameron VA, et al. Circulating miR-323-3p and miR-652: candidate markers for the presence and progression of acute coronary syndromes. Int J Cardiol 2014;176:375-85. [PubMed]
- He F, Lv P, Zhao X, et al. Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Mol Cell Biochem 2014;394:137-44. [PubMed]
- Meder B, Keller A, Vogel B, et al. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res Cardiol 2011;106:13-23. [PubMed]
- Vogel B, Keller A, Frese KS, et al. Refining diagnostic microRNA signatures by whole-miRNome kinetic analysis in acute myocardial infarction. Clin Chem 2013;59:410-8. [PubMed]
- Olson EN. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Sci Transl Med 2014;6:239ps3.
- Wang Z. The guideline of the design and validation of MiRNA mimics. Methods Mol Biol 2011;676:211-23. [PubMed]
- Caroli A, Cardillo MT, Galea R, et al. Potential therapeutic role of microRNAs in ischemic heart disease. J Cardiol 2013;61:315-20. [PubMed]
- Krützfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005;438:685-9. [PubMed]
- Oliveira-Carvalho V, Carvalho VO, Silva MM, et al. MicroRNAs: a new paradigm in the treatment and diagnosis of heart failure? Arq Bras Cardiol 2012;98:362-9. [PubMed]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007;4:721-6. [PubMed]
- Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 2007;39:1033-7. [PubMed]
- Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol 2010;20:R858-61. [PubMed]
- Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. Rna 2010;16:2043-50. [PubMed]
- Xiao J, Yang B, Lin H, et al. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination on the pacemaker channel genes HCN2 and HCN4. J Cell Physiol 2007;212:285-92. [PubMed]
- Feng B, Cao Y, Chen S, et al. Reprint of: miRNA-1 regulates endothelin-1 in diabetes. Life Sci 2014;118:275-80. [PubMed]
- Yin C, Salloum FN, Kukreja RC. A novel role of microRNA in late preconditioning: upregulation of endothelial nitric oxide synthase and heat shock protein 70. Circ Res 2009;104:572-5. [PubMed]
- Huang F, Li ML, Fang ZF, et al. Overexpression of MicroRNA-1 improves the efficacy of mesenchymal stem cell transplantation after myocardial infarction. Cardiology 2013;125:18-30. [PubMed]
- Ren XP, Wu J, Wang X, et al. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation 2009;119:2357-66. [PubMed]
- Ye Y, Hu Z, Lin Y, et al. Downregulation of microRNA-29 by antisense inhibitors and a PPAR-gamma agonist protects against myocardial ischaemia-reperfusion injury. Cardiovasc Res 2010;87:535-44. [PubMed]
- Becker N, Lockwood CM. Pre-analytical variables in miRNA analysis. Clin Biochem 2013;46:861-8. [PubMed]
- Monleau M, Bonnel S, Gostan T, et al. Comparison of different extraction techniques to profile microRNAs from human sera and peripheral blood mononuclear cells. BMC Genomics 2014;15:395. [PubMed]
- Git A, Dvinge H, Salmon-Divon M, et al. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. Rna 2010;16:991-1006. [PubMed]
- Meyer SU, Kaiser S, Wagner C, et al. Profound effect of profiling platform and normalization strategy on detection of differentially expressed microRNAs--a comparative study. PLoS One 2012;7:e38946. [PubMed]
- Hardikar AA, Farr RJ, Joglekar MV. Circulating microRNAs: understanding the limits for quantitative measurement by real-time PCR. J Am Heart Assoc 2014;3:e000792. [PubMed]
- Zampetaki A, Mayr M. Analytical challenges and technical limitations in assessing circulating miRNAs. Thromb Haemost 2012;108:592-8. [PubMed]
- Huang X, Liang M, Dittmar R, et al. Extracellular microRNAs in urologic malignancies: chances and challenges. Int J Mol Sci 2013;14:14785-99. [PubMed]
- Kaudewitz D, Lee R, Willeit P, et al. Impact of intravenous heparin on quantification of circulating microRNAs in patients with coronary artery disease. Thromb Haemost 2013;110:609-15. [PubMed]
- Boeckel JN, Thomé CE, Leistner D, et al. Heparin selectively affects the quantification of microRNAs in human blood samples. Clin Chem 2013;59:1125-7. [PubMed]
- Siemelink MA, Zeller T. Biomarkers of coronary artery disease: the promise of the transcriptome. Curr Cardiol Rep 2014;16:513. [PubMed]
- Zeller T, Hughes M, Tuovinen T, et al. BiomarCaRE: rationale and design of the European BiomarCaRE project including 300,000 participants from 13 European countries. Eur J Epidemiol 2014;29:777-90. [PubMed]
- Skroblin P, Mayr M. “Going Long”: Long Non-Coding RNAs as Biomarkers. Circ Res 2014;115:607-9. [PubMed]
- Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 2013;20:300-7. [PubMed]
- Vausort M, Wagner DR, Devaux Y. Long Noncoding RNAs in Patients With Acute Myocardial Infarction. Circ Res 2014;115:668-77. [PubMed]


