The cardiac hypoxic niche: emerging role of hypoxic microenvironment in cardiac progenitors
Hifs and hypoxia response
In aerobic organisms, cells require the supply of oxygen (O2) for generating intracellular ATP through oxidative phosphorylation. O2 availability profoundly affects almost all biological processes, and therefore, the response to hypoxia, is critical for all organisms. During hypoxia, cells activate a number of adaptive physiological responses, which directly influence metabolism, redox homeostasis, vascular remodeling etc. Hypoxia inducible factors (Hifs) are key modulators of the hypoxia-induced transcriptional program designed to counteract the reduced O2 availability at both cellular and systemic levels (1). Hifs are heterodimeric factors consisting of an O2-regulated alpha subunit and constitutively expressed beta subunit. Three isoforms of Hifα have been identified in mammals. Hif-1α and Hif-2α are highly related in their amino acid structures, and molecular and biological roles of them are well characterized. Hif-1α is expressed ubiquitously, but Hif-2α and Hif-3α, in contrast, are selectively expressed in certain tissues, including vascular endothelial cells (2) and peritubular interstitial cells in the kidney (3). Hif-1α expression induces a switch from the oxygen-dependent oxidative metabolism in the mitochondria to anaerobic glycolytic metabolism in the cytoplasm, whereas Hif-2α is believed to be a master regulator of oxidant stress response (4). The function of Hif-3α is not entirely understood, but it is known to possess a variety of splicing isoforms and some of them act as dominant negative forms of Hif-1α and Hif-2α (5-10).
Regulation of Hif activity by O2 tension has been demonstrated at transcriptional, translational and post-transcriptional levels. O2-dependent control of Hifα protein stability is one of the classic and best characterized regulatory mechanism (Figure 1A). Under normal O2 tension, or normoxic conditions, Hifα subunits are hydroxylated at conserved proline residues in the oxygen dependent degradation (ODD) domain (9). These modifications are mediated by three prolyl hydroxylase domain-containing enzymes (PHD1-3), which require O2 for their enzymatic activity (9). Prolyl-hydroxylated Hifα is then recognized by the E3-ubiquitin-ligase, von Hippel-Lindau (VHL) complex and degraded by the ubiquitin-mediated proteasome pathway. Under hypoxic conditions, Hifα prolyl hydroxylation is suppressed due to the lack of oxygen as a substrate of hydroxylation reaction and consequently Hifα proteins stabilization.
Recently more factors which regulate Hif protein activity have been reported, such as factor-inhibiting Hif-1α (FIH1) an asparaginyl hydroxylase, histone deacetylases Sirtuins, several oncogenes and tumor suppressor factors, as well as a number of transcriptional regulators [reviewed by Majmundar et al. (11) and Semenza (12)], suggesting more complex and precise regulation of Hif complexes in several different cellular contexts (Figure 1B).
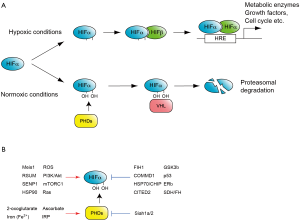 Figure 1. O dependent HIF regulation. A. One of the classic and well-studied mechanisms regulating HIF activity by O. Under normoxic conditions HIFα is hydroxylated by prolyl hydroxylase domain-containing enzymes (PHDs), and recognized by the E3-ubiquitin-ligase, von Hippel-Lindau (VHL) to be subjected to proteasome-mediated degradation. Under hypoxic conditions HIFα is stable and together with constitutive HIFα, binds to hypoxia response element (HRE) to activate hundreds of target genes; B. Factors activate or repress HIF activity. Note that these factors modulate HIF at several different levels. For example Meis1 activates the transcription of HIFα mRNA, whereas RSUME and SEMP1 mediate SUMOylation of HIFα proteins
Figure 1. O dependent HIF regulation. A. One of the classic and well-studied mechanisms regulating HIF activity by O. Under normoxic conditions HIFα is hydroxylated by prolyl hydroxylase domain-containing enzymes (PHDs), and recognized by the E3-ubiquitin-ligase, von Hippel-Lindau (VHL) to be subjected to proteasome-mediated degradation. Under hypoxic conditions HIFα is stable and together with constitutive HIFα, binds to hypoxia response element (HRE) to activate hundreds of target genes; B. Factors activate or repress HIF activity. Note that these factors modulate HIF at several different levels. For example Meis1 activates the transcription of HIFα mRNA, whereas RSUME and SEMP1 mediate SUMOylation of HIFα proteins
During hypoxia, Hifα is stabilized and forms heterodimers with Hifβ, which causes conformational change in Hif-α that allows the HIF heterodimer to bind to hypoxia response elements (HREs) (13) scattered throughout the genome and activate the transcription of several hundreds of target genes (14,15). Typical examples of the target genes of Hif heterodimers include erythropoietin (16,17) and vascular endothelial growth factor receptor (18,19), which activate erythropoiesis and angiogenesis, respectively. Hif complexes also regulates genes that activate glycolysis, which include glucose transporters [such glucose transporter 1 (GLUT1)], glycolytic regulatory enzymes like 6-phospho-2-kinase/fructose 2, 6-biphosphatase (PFKFB1-4), hexokinase II (HKII) and lactate dehydrogenase A (LDHA), as well as activating genes which diminish mitochondrial oxidative metabolism, including pyruvate dehydrogenase kinase 1 (PDK1) [reviewed by Iyer et al. (20), Bartrons and Caro (21), and Wheaton and Chandel (22)]. The net result is a metabolic switch from mitochondrial phosphorylation to glycolysis. These Hif complex mediated transcriptional programs regulate many biological processes including embryonic development, tumor progression and also stem cell properties such as maintenance, self-renewal and differentiation.
Hypoxia signaling in adult stem cells
Hematopoietic stem cells (HSCs), which have capacities for both self-renewal and multi-lineage differentiation, continue to replenish all blood cells throughout the entire life span of an organism (23,24). The bone marrow microenvironment which house HSCs, known as niches, provide HSCs with regulatory signals essential for their maintenance, proliferation and differentiation. One of the hallmarks of the HSC niche is its low oxygen tension, hence the term “hypoxic niche” (25). A number of studies revealed that this hypoxic environment is required for HSC quiescence. Moreover, colony-forming ability and transplantation capacity increases when bone marrow cells are cultured under low oxygen tension (26-28). Therefore, this low oxygen environment is not only tolerated by HSCs, but is also essential to maintain their function (26,29,30). Our group, as well as others, recently reported that Hif-1α mRNA and protein are highly expressed in LT-HSCs where it plays a crucial role in role in the maintenance of HSC quiescence and stress resistance (31-33).
Since HSCs are sustained for a long time, they have to evolve mechanisms to diminish, and resist many stressors including oxidative stress. Our group revealed that LT-HSCs utilize cytoplasmic glycolysis, instead of mitochondrial respiration for their energy production (32), thereby minimizing mitochondrial derived reactive oxygen species (ROS). We showed that HSC-specific deletion of Hif-1α results in increased rates of mitochondrial respiration and decreased glycolytic flux (33). Interestingly, Hif-1α appears to be regulated at multiple levels in HSCs. We recently reported that homeodomain transcription factor Meis1 plays an essential role in the maintenance of LT-HSC through transcriptional activation for Hif-1α (32,33) (Figure 2). Meis1 also regulates Hif-2α transcription, where loss of Meis1 results in downregulation of Hif-2α levels, and increased ROS levels. Intriguingly, systemic administration of ROS scavengers rescues the Meis1-/- phenotype in HSCs. These results strongly support the central role of hypoxia signaling and redox regulation in HSC maintenance and survival (32,33).
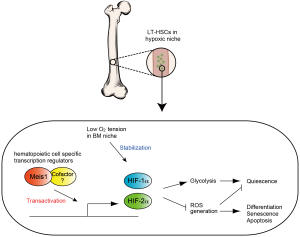 Figure 2. In LT-HCSs, Meis1 (and its cofactor) upregulate both HIF-1α and HIF-2α. This transcriptional activation is essential for maintaining the glycolytic metabolic phenotype and preventing ROS mediated senescence and apoptosis
Figure 2. In LT-HCSs, Meis1 (and its cofactor) upregulate both HIF-1α and HIF-2α. This transcriptional activation is essential for maintaining the glycolytic metabolic phenotype and preventing ROS mediated senescence and apoptosis
In addition to the hematopoietic system, hypoxia and Hif genes also play important roles in other adult stem cell populations. The subventricular zone of the hippocampus, known as a neural stem cell (NSC) niche, shares low oxygen tension properties with the hypoxic HSC niche (34), where loss of Hif-1α decreases NSC proliferation, differentiation and neural maturation (34). Moreover, Hifs are upregulated in several cancer stem cells and play roles in their survival and differentiation [reviewed by Semenza (12), Kobayashi and Suda (35), Heddleston et al. (36), and Li and Rich (37)]. For example, Hif-1α and Hif-2α is expressed in glioma stem cells and are essential for their self-renewal and tumorigenic capacity (38,39). In addition, the Hif-1α inhibitor echinomycin eradicates lymphoma and acute myeloid leukemia by eliminating cancer stem cells (40). Thus, hypoxia and Hif-1α upregulation appears to be a features shared by various types of stem cells and an important regulator of their stem cell properties.
Tissue specific cardiac progenitors
It has been long thought that cardiomyocyte turnover does not occur in the adult mammalian hearts, because myocardial injury invariably results in irreversible scar formation (41-43). However, recent evidence suggests that the adult mammalian heart does in fact undergo some degree of cardiomyocyte renewal during normal aging (44-48) and disease (45,46,48-50). Unfortunately, this modest myocyte renewal is insufficient for restoration of contractile function following injury. In contrast, some species of fish (51,52) and amphibians (53-56), as well as fetal (57) and neonatal mammals (58) have a remarkable ability to regenerate damaged myocardium, which is mainly achieved by proliferation of pre-existing cardiomyocyte (58-61). Interestingly, current evidence suggests that the adult mammalian heart derives the small number of new cardiomyocytes from an unknown stem or progenitor cell source (62).
Several reports indicate that the adult rodent and human myocardium harbors populations of stem/progenitor cells, which appear to have the potential to generate cardiomyocytes in vitro (63-65). These cells are potentially an attractive source for cardiac repair and therefore, over the past decade, extensive studies have examined their use for cardiac regeneration [reviewed by Laflamme and Murry (49), Hansson et al. (66), and Choi and Poss (67)]. In mice, cells expressing the stem cell factor receptor c-kit(68), stem cell antigen-1 [Sca-1, (69)], the transcription factor islet-1 [Isl-1, (70)], and side population (SP) cells (71) have been identified as cardiac resident progenitor cells due to their ability to acquire a cardiomyocyte lineage in vivo. Moreover, progenitor cells in the postnatal human hearts have been also identified by the expression of c-kit, Sca-1, Isl-1 (72-77) or SP profile (78). Even though SP cells, c-kit+ cells, Sca-1+ cells and Isl1+ cells all have a capacity to differentiate into smooth muscle cells, endothelial cells and cardiomyocyte, they are clearly distinct cell population in several aspects. For example, Isl1 positive progenitor cells do not express c-kit as oppose to SP, Sca-1 positive and c-kit positive progenitor cells [reviewed by Guan and Hasenfuss (79), and Bollini et al. (80)]. In addition, their localization within the heart appears to be distinct, where SP cells, Sca-1+ cells and c-kit+ cells are mostly enriched in the atria (73,74), Isl1+ cells are found widely in ventricles and atria (70). It is therefore critical to determine which lineages these cells contribute to during normal cardiac homeostasis, ageing and after injury. However, definitive genetic fate mapping studies are still underway to conclusively determine which of these diverse cell populations are true cardiac stem or progenitor cells.
The epicardium as a source of cardiac progenitor cells
Another type of resident cardiac progenitors has been identified based on anatomical localization to the epicardium, and expression of epicardial markers. In zebrafish, the epicardium is activated within 1-2 days after injury (60,81,82), where it proliferates, expresses embryonic epicardial genes, and undergoes epithelial-mesenchymal transition (82-84). The adult mammalian epicardial cells are activated similarly by myocardial infarction and/or Thymosin β4 stimulation (85), where they proliferate, start to express embryonic epicardial genes (86), and secrete paracrine factors that modified the myocardial injury response (83,87-90).
In the past few years, increasing evidences suggested that the epicardium also contains distinct progenitor cells during normal development and upon injury (reviewed by Schlueter and Brand (91,92). Epicardial derived cells contribute to cardiac fibroblast, vascular smooth muscle cells and cardiomyocytes during embryogenesis by lineage tracing with WT1-Cre (93) and Tbx18-Cre (94) lines [although the epicardial origin of cardiomyocyte remains controversial because it was revealed that the Tb ×18 is not only expressed in epicardium, but also in developing cardiomyocytes (95)]. Moreover, Chong et al. showed that the adult epicardium contains multipotent stem cells, which have the capacity to differentiate into cardiomyocytes, vascular endothelial and smooth muscle cells (96). Moreover, taking advantage of the upregulation of WT1 in epicardium in the adult heart upon injury, Zhou et al. revealed by lineage tracing that adult epicardium-derived cells contribute to fibroblasts and perivascular smooth muscle cells, without contribution to the cardiomyocyte pool (83), except after priming with thymosin beta 4 (85). These studies support the role of epicardial cells as cardiac progenitors, although fate-mapping studies that unequivocally demonstrate that the epicardium is the source of turnover of cardiomyocytes are still lacking.
Hypoxic cardiac progenitor cells
These studies reviewed above identify the epicardium as a potential source of cardiac progenitors, however whether the epicardium is the true cardiac progenitor/stem cell niche, and whether it shares the hypoxic properties with other stem cell niches in the adult organism is not clear. To answer this question, we recently sought to identify hypoxic regions in the uninjured adult mammalian heart, and we found that the epicardium and subepicardium represent the cardiac hypoxic niche (97). The epicardial and subepicardium (within three-cell layer from the epicardium) have the lowest capillary density across the ventricle, with over 50% of epicardial cells and less than 10% of the subepicardial cardiomyocytes express Hif-1α protein. Moreover, non cardiomyocytes isolated from this region and expanded in culture, are clonogenic, self-renewing, express cardiac progenitor and epicardial markers, and are capable of acquiring different cardiac lineages in vitro including endothelium, smooth muscle and cardiomyocyte lineages. These results provide proof that the epicardium and subepicrdium represent the hypoxic niche of adult heart which, like other hypoxic niches in the adult organism, houses a population of cardiac progenitor cells (Figure 3).
 Figure 3. Cardiac hypoxic niche in epicardium and subepicardium. The hypoxic microenvironment houses metabolically distinct population of glycolytic progenitor cells (GCPs). Epicardial GCPs isolated based on low mitochondrial content are c-kit-; Sca-1-; Nk×2.5+ and can differentiate into endothelial cells, smooth muscle cells and cardiomyocytes
Figure 3. Cardiac hypoxic niche in epicardium and subepicardium. The hypoxic microenvironment houses metabolically distinct population of glycolytic progenitor cells (GCPs). Epicardial GCPs isolated based on low mitochondrial content are c-kit-; Sca-1-; Nk×2.5+ and can differentiate into endothelial cells, smooth muscle cells and cardiomyocytes
In support of the link between the metabolic phenotype and stem or progenitor cell phenotype, we found that these hypoxic cardiac niche cells mainly rely on cytoplasmic glycolysis, rather than mitochondrial oxidative phosphorylation (and therefore we named them glycolytic cardiac progenitors, or GCPs). Not surprisingly, knockdown of Hif-1α in these cells resulted in metabolic shift from glycolysis to mitochondrial oxidative phosphorylation, which was associated with decreased in their rate of proliferation and spontaneous differentiation. These findings suggest that similar to LT-HSCs, Hif-1α is required for maintaining self-renewing stem/progenitor cells within the hypoxic niche by regulating their glycolytic phenotype (97).
A number of questions regarding hypoxic cardiac progenitor cells still remain unanswered. For example, do they contribute to the cardiomyocyte turnover during normal aging and after myocardial injury? To answer this question conclusively, fate-mapping studies are necessary. However lineage tracing of GCPs is currently not technically feasible, in part because there are no known specific marker of GCPs or of cardiac hypoxic niche cells. In addition, what is the relationship between the hypoxic response (for example which occurs after myocardial infarction) and proliferation, migration and differentiation of GCPs in vivo? It is known that high ROS levels force other types of stem cells (such as HSCs) to exit from quiescence, spontaneously differentiate, and lose their self-renewal capacity (98-100). ROS levels markedly increase in the myocardium following an acute myocardial infarction (101,102), at the time of ischemia/reperfusion (103-105) and in chronic heart failure (106-108), where they play a major role in mediating cardiomyocyte injury and death [reviewed by (109-111)]. Taking the similarities between HSCs and GCPs into consideration, it is important to determine how an in vivo ROS-rich environment would affect GCPs, and whether this plays a role in the deleterious effects of ROS on the myocardium. Another important question is the potential mechanism of migration of epicardial/subepircardial cells toward the injured myocardium. It is thought that epicardial/subepicardial cells can migrate into myocardial wall and differentiate into cardiomyocyte (85), however, factors that control their migration remain still unknown. One possible mechanism is the hypoxia itself that occurs as the result of injury (112,113). Hypoxia is known to promote stem cell recruitment, for example HSC homing into hypoxic bone marrow niche, via Hif-1α mediated production of chemokine SDF-1/CXCL12 [reviewed by Shiozawa et al. (114), Kavanagh and Kalia (115), Suárez-Álvarez et al. (116), and Schulz et al. (117)]. Chemokines including SDF-1 are also upregulated in the heart after acute myocardial infarction and attract various progenitor cell populations, which possess cardioprotective properties [reviewed by Ghadge et al. (118), and Smart and Riley (119)]. Moreover, in zebrafish SDF-1/CXCL12 is reported to regulate cardiomyocyte migration after injury (120). Therefore, it is plausible that hypoxia plays a dual role in maintenance as well as recruitment of epicardial cells after injury. However, a full understanding of the mechanisms that govern homing and migration of epicardial cells is still lacking.
Concluding remarks
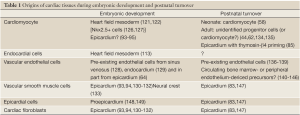
One important question that presents itself is whether there is a true cardiac stem cell in the adult mammalian heart. While other adult tissues, like the hematopoietic system, certainly have true stem cells, it is unclear if this is the case in the heart, especially if one takes clues from cellular origin of the heart during development. A true adult heart stem cell, from the differentiation perspective, would have to give rise to all cells within the heart including endothelial cells, smooth muscle cells, fibroblasts and cardiomyocytes, however these cells are actually quite distinct developmentally. During embryonic heart formation, two different major sources in splanchnic mesoderm provide cardiac progenitor cells: one is heart field mesoderm, and the other is proepicardium [reviewed by Dyer and Kirby (121); Abu-Issa and Kirby (122); Lie-Venema et al. (123) and Ishii et al. (124)]. Heart field mesoderm mainly contributes to cardiomyocytes (Nk ×2.5+ cells), endocardium and vascular endothelium (Flk1+ cells) (125-127). However, most of the cardiac vascular endothelial cells are descendants of pre-existing endothelial cells in sinus venosus (128) with some contribution from the endocardium (129) from which cells sprout out and migrate within the myocardial wall. On the other hand, the proepicardium gives rise to epicardial cells, cardiac fibroblasts, part of vascular endothelium and vascular smooth muscle cells (93,94,130-132) [coronary smooth muscle cells are also derived from cardiac neural crest (133)]. Therefore, while the concept of a true cardiac stem cell is highly appealing, there is no definitive proof (by genetic fate mapping) to indicate that this type of cell exists and contributes to cellular turnover as in case of HCSs. In fact, the current evidence suggests that turnover of cells in the post-natal heart resembles that during development (Table 1). For example, fibroblasts and vascular smooth muscle cells are derived from the epicardium (93,94), vascular endothelial cells are descendants of proliferating and migrating endothelial cells originated from adjacent endothelium (136-139) and/or circulating progenitor cells of bone marrow or peripheral endothelium (140-143) (although recently it has been questioned that these circulating progenitor cells really contribute to newly generated endothelial cells after the injury (144-146). Moreover, post-natal cardiomyocytes appear to be derived from pre-existing cardiomyocytes (58,134,135), the epicardium [only after thymosin beta 4 priming (85)] or unknown progenitor cells (44,62). It is perhaps intriguing that these cell lineages, which replenish the adult heart, appear to recapitulate their developmental origin, and very rarely cross the boundary of major developmental lineage sources.
Full table
It is important to remember that even if there is currently no irrefutable evidence of a “true” adult cardiac stem cell that carries out the function of replenishing all cell lineages in the heart during ageing and disease, this does not mean that many of the cell populations described above have no therapeutic value. On the contrary, mounting evidence suggests that cell therapy may in fact induce a measurable regenerative response in the adult myocardium, unrelated to their transdifferentiation capacity. An elegant example of a regenerative role of C-Kit cells was recently demonstrated by the Lee lab, where they clearly showed an increase in the number of newly formed cardiomyocytes, and improved systolic function after injection of C-Kit cells, although these cells were no longer present in the heart (150).
In this review we summarized recent findings that highlight the emerging role of hypoxia signaling in stem cell niches, including the newly identified cardiac hypoxic niche. We propose that better characterizing of the epicardial hypoxic niche cells, in terms of their lineage differentiation potential, and the role of hypoxia signaling in regulating their function, are important targets for modulating the capability of resident cardiac stem/progenitor cells for therapeutic applications.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Lahiri S, Roy A, Baby SM, et al. Oxygen sensing in the body. Prog Biophys Mol Biol 2006;91:249-86.
- Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer 2008;8:967-75.
- Rosenberger C, Mandriota S, Jurgensen JS, et al. Expression of hypoxia-inducible factor-1alpha and -2alpha in hypoxic and ischemic rat kidneys. J Am Soc Nephrol 2002;13:1721-32.
- Scortegagna M, Ding K, Oktay Y, et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1-/- mice. Nat Genet 2003;35:331-40.
- Gu YZ, Moran SM, Hogenesch JB, et al. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expression 1998;7:205-13.
- Makino Y, Cao R, Svensson K, et al. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature 2001;414:550-4.
- Jang MS, Park JE, Lee JA, et al. Binding and regulation of hypoxia-inducible factor-1 by the inhibitory PAS proteins. Biochem Biophys Res Commun 2005;337:209-15.
- Maynard MA, Evans AJ, Hosomi T, et al. Human HIF-3alpha4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. Faseb J 2005;19:1396-406.
- Kaelin WG Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 2008;30:393-402.
- Heikkilä M, Pasanen A, Kivirikko KI, et al. Roles of the human hypoxia-inducible factor (HIF)-3alpha variants in the hypoxia response. Cell Mol Life Sci 2011;68:3885-901.
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell 2010;40:294-309.
- Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci 2012;33:207-14.
- Kallio PJ, Pongratz I, Gradin K, et al. Activation of hypoxia-inducible factor 1alpha: posttranscriptional regulation and conformational change by recruitment of the Arnt transcription factor. Proc Natl Acad Sci U S A 1997;94:5667-72.
- Xia X, Lemieux ME, Li W, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A 2009;106:4260-5.
- Schödel J, Oikonomopoulos S, Ragoussis J, et al. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 2011;117:e207-17.
- Köchling J, Curtin PT, Madan A. Regulation of human erythropoietin gene induction by upstream flanking sequences in transgenic mice. Br J Haematol 1998;103:960-8.
- Warnecke C, Zaborowska Z, Kurreck J, et al. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. Faseb J 2004;18:1462-4.
- Huang LE, Arany Z, Livingston DM, et al. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol. Chem 1996;271:32253-9.
- Sutter CH, Laughner E, Semenza GL. Hypoxia-inducible factor 1alpha protein expression is controlled by oxygen-regulated ubiquitination that is disrupted by deletions and missense mutations. Proc Natl Acad Sci U S A 2000;97:4748-53.
- Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev 1998;12:149-62.
- Bartrons R, Caro J. Hypoxia, glucose metabolism and the Warburg’s effect. J Bioenerg Biomembr 2007;39:223-9.
- Wheaton WW, Chandel NS. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am J Physiol Cell Physiol 2011;300:C385-93.
- Abramson S, Miller RG, Phillips RA. The identification in adult bone marrow of pluripotent and restricted stem cells of the myeloid and lymphoid systems. J Exp Med 1977;145:1567-79.
- Jordan CT, McKearn JP, Lemischka IR. Cellular and developmental properties of fetal hematopoietic stem cells. Cell 1990;61:953-63.
- Eliasson P, Jonsson JI. The hematopoietic stem cell niche: low in oxygen but a nice place to be. J Cell Physiol 2010;222:17-22.
- Cipolleschi MG, Dello Sbarba P, Olivotto M. The role of hypoxia in the maintenance of hematopoietic stem cells. Blood 1993;82:2031-7.
- Danet GH, Pan Y, Luongo JL, et al. Expansion of human SCID-repopulating cells under hypoxic conditions. J Clin Invest 2003;112:126-35.
- Ivanovic Z, Hermitte F, Brunet de la Grange P, et al. Simultaneous maintenance of human cord blood SCID-repopulating cells and expansion of committed progenitors at low O2 concentration (3%). Stem Cells 2004;22:716-24.
- Studer L, Csete M, Lee SH, et al. Enhanced proliferation, survival, and dopaminergic differentiation of CNS precursors in lowered oxygen. J Neurosci 2000;20:7377-83.
- Ivanović Z, Dello Sbarba P, Trimoreau F, et al. Primitive human HPCs are better maintained and expanded in vitro at 1 percent oxygen than at 20 percent. Transfusion 2000;40:1482-8.
- Takubo K, Goda N, Yamada W, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 2010;7:391-402.
- Simsek T, Kocabas F, Zheng J, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 2010;7:380-90.
- Kocabas F, Zheng J, Thet S, et al. Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood 2012;120:4963-72.
- Mazumdar J, O’Brien WT, Johnson RS, et al. O2 regulates stem cells through Wnt/beta-catenin signalling. Nat Cell Biol 2010;12:1007-13.
- Kobayashi CI, Suda T. Regulation of reactive oxygen species in stem cells and cancer stem cells. J Cell Physiol 2012;227:421-30.
- Heddleston JM, Li Z, Lathia JD, et al. Hypoxia inducible factors in cancer stem cells. Br J Cancer 2010;102:789-95.
- Li Z, Rich JN. Hypoxia and hypoxia inducible factors in cancer stem cell maintenance. Curr Top Microbiol Immunol 2010;345:21-30.
- Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 2009;15:501-13.
- Soeda A, Park M, Lee D, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene 2009;28:3949-59.
- Wang Y, Liu Y, Malek SN, et al. Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell Stem Cell 2011;8:399-411.
- Murry CE, Reinecke H, Pabon LM. Regeneration gaps: observations on stem cells and cardiac repair. J Am Coll Cardiol 2006;47:1777-85.
- Laflamme MA, Murry CE. Regenerating the heart. Nat Biotechnol 2005;23:845-56.
- Whelan RS, Kaplinskiy V, Kitsis RN. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu Rev Physiol 2010;72:19-44.
- Bergmann O, Bhardwaj RD, Bernard S, et al. Evidence for cardiomyocyte renewal in humans. Science 2009;324:98-102.
- Soonpaa MH, Field LJ. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am J Physiol 1997;272:H220-6.
- Soonpaa MH, Field LJ. Survey of studies examining mammalian cardiomyocyte DNA synthesis. Circ Res 1998;83:15-26.
- Kajstura J, Urbanek K, Perl S, et al. Cardiomyogenesis in the adult human heart. Circ Res 2010;107:305-15.
- Kajstura J, Rota M, Cappetta D, et al. Cardiomyogenesis in the aging and failing human heart. Circulation 2012;126:1869-81.
- Laflamme MA, Murry CE. Heart regeneration. Nature 2011;473:326-35.
- Laflamme MA, Myerson D, Saffitz JE, et al. Evidence for cardiomyocyte repopulation by extracardiac progenitors in transplanted human hearts. Circ Res 2002;90:634-40.
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science 2002;298:2188-90.
- Raya A, Koth CM, Buscher D, et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci U S A 2003;100:11889-95.
- Rumyantsev PP. Post-injury DNA synthesis, mitosis and ultrastructural reorganization of adult frog cardiac myocytes. An electron microscopic-autoradiographic study. Z Zellforsch Mikrosk Anat 1973;139:431-50.
- Flink IL. Cell cycle reentry of ventricular and atrial cardiomyocytes and cells within the epicardium following amputation of the ventricular apex in the axolotl, Amblystoma mexicanum: confocal microscopic immunofluorescent image analysis of bromodeoxyuridine-labeled nuclei. Anat Embryol (Berl) 2002;205:235-44.
- Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. J Exp Zool 1974;187:249-53.
- Witman N, Murtuza B, Davis B, et al. Recapitulation of developmental cardiogenesis governs the morphological and functional regeneration of adult newt hearts following injury. Dev Biol 2011;354:67-76.
- Drenckhahn JD, Schwarz QP, Gray S, et al. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev Cell 2008;15:521-33.
- Porrello ER, Mahmoud AI, Simpson E, et al. Transient regenerative potential of the neonatal mouse heart. Science 2011;331:1078-80.
- Jopling C, Sleep E, Raya M, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010;464:606-9.
- Kikuchi K, Holdway JE, Werdich AA, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 2010;464:601-5.
- Wang J, Panakova D, Kikuchi K, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 2011;138:3421-30.
- Hsieh PC, Segers VF, Davis ME, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med 2007;13:970-4.
- Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell 2008;2:320-31.
- Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell 2008;132:537-43.
- Anversa P, Nadal-Ginard B. Myocyte renewal and ventricular remodelling. Nature 2002;415:240-3.
- Hansson EM, Lindsay ME, Chien KR. Regeneration next: toward heart stem cell therapeutics. Cell Stem Cell 2009;5:364-77.
- Choi WY, Poss KD. Cardiac regeneration. Curr Top Dev Biol 2012;100:319-44.
- Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003;114:763-76.
- Oh H, Bradfute SB, Gallardo TD, et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci U S A 2003;100:12313-8.
- Laugwitz KL, Moretti A, Lam J, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 2005;433:647-53.
- Hierlihy AM, Seale P, Lobe CG, et al. The post-natal heart contains a myocardial stem cell population. FEBS Lett 2002;530:239-43.
- Urbanek K, Torella D, Sheikh F, et al. Myocardial regeneration by activation of multipotent cardiac stem cells in ischemic heart failure. Proc Natl Acad Sci U S A 2005;102:8692-7.
- Itzhaki-Alfia A, Leor J, Raanani E, et al. Patient characteristics and cell source determine the number of isolated human cardiac progenitor cells. Circulation 2009;120:2559-66.
- van Vliet P, Roccio M, Smits AM, et al. Progenitor cells isolated from the human heart: a potential cell source for regenerative therapy. Neth Heart J 2008;16:163-9.
- Messina E, De Angelis L, Frati G, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res 2004;95:911-21.
- Chimenti I, Smith RR, Li TS, et al. Relative roles of direct regeneration versus paracrine effects of human cardiosphere-derived cells transplanted into infarcted mice. Circ Res 2010;106:971-80.
- Bearzi C, Rota M, Hosoda T, et al. Human cardiac stem cells. Proc Natl Acad Sci U S A 2007;104:14068-73.
- Emmert MY, Emmert LS, Martens A, et al. Higher frequencies of BCRP+ cardiac resident cells in ischaemic human myocardium. Eur Heart J 2012. [Epub ahead of print].
- Guan K, Hasenfuss G. Cardiac resident progenitor cells: evidence and functional significance. Eur Heart J 2012. [Epub ahead of print].
- Bollini S, Smart N, Riley PR. Resident cardiac progenitor cells: at the heart of regeneration. J Mol Cell Cardiol 2011;50:296-303.
- Kikuchi K, Holdway JE, Major RJ, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell 2011;20:397-404.
- Lepilina A, Coon AN, Kikuchi K, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 2006;127:607-19.
- Zhou B, Honor LB, He H, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest 2011;121:1894-904.
- Kikuchi K, Gupta V, Wang J, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 2011;138:2895-902.
- Smart N, Bollini S, Dube KN, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature 2011;474:640-4.
- Merki E, Zamora M, Raya A, et al. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A 2005;102:18455-60.
- Russell JL, Goetsch SC, Gaiano NR. A dynamic notch injury response activates epicardium and contributes to fibrosis repair. Circ Res 2011;108:51-9.
- Winter EM, Grauss RW, Hogers B, et al. Preservation of left ventricular function and attenuation of remodeling after transplantation of human epicardium-derived cells into the infarcted mouse heart. Circulation 2007;116:917-27.
- Duan J, Gherghe C, Liu D, et al. Wnt1/betacatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. Embo J 2012;31:429-42.
- Bock-Marquette I, Saxena A, White MD, et al. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature 2004;432:466-72.
- Schlueter J, Brand T. Epicardial progenitor cells in cardiac development and regeneration. J Cardiovasc Transl Res 2012;5:641-53.
- Gittenberger-de Groot AC, Winter EM, Poelmann RE. Epicardium-derived cells (EPDCs) in development, cardiac disease and repair of ischemia. J Cell Mol Med 2010;14:1056-60.
- Zhou B, Ma Q, Rajagopal S, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 2008;454:109-13.
- Cai CL, Martin JC, Sun Y, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature 2008;454:104-8.
- Christoffels VM, Grieskamp T, Norden J, et al. Tbx18 and the fate of epicardial progenitors. Nature 2009;458:E8-9; discussion E9-10.
- Chong JJ, Chandrakanthan V, Xaymardan M, et al. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell 2011;9:527-40.
- Kocabas F, Mahmoud AI, Sosic D, et al. The hypoxic epicardial and subepicardial microenvironment. J Cardiovasc Transl Res 2012;5:654-65.
- Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood 2007;110:3056-63.
- Ito K, Hirao A, Arai F, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med 2006;12:446-51.
- Tothova Z, Kollipara R, Huntly BJ, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 2007;128:325-39.
- Jennings RB, Reimer KA. Acute myocardial ischemia: effects of reperfusion with arterial blood. Artif Cells Blood Substit Immobil Biotechnol 1994;22:253-78.
- Frangogiannis NG, Smith CW, Entman ML. The inflammatory response in myocardial infarction. Cardiovasc Res 2002;53:31-47.
- Becker LB, vanden Hoek TL, Shao ZH, et al. Generation of superoxide in cardiomyocytes during ischemia before reperfusion. Am J Physiol 1999;277:H2240-6.
- Eaton P, Li JM, Hearse DJ, et al. Formation of 4-hydroxy-2-nonenal-modified proteins in ischemic rat heart. Am J Physiol 1999;276:H935-43.
- Kevin LG, Camara AK, Riess ML, et al. Ischemic preconditioning alters real-time measure of O2 radicals in intact hearts with ischemia and reperfusion. Am J Physiol Heart Circ Physiol 2003;284:H566-74.
- McMurray J, Chopra M, Abdullah I, et al. Evidence of oxidative stress in chronic heart failure in humans. Eur Heart J 1993;14:1493-8.
- Dhalla NS, Temsah RM, Netticadan T. Role of oxidative stress in cardiovascular diseases. J Hypertens 2000;18:655-73.
- Li YL, Gao L, Zucker IH, et al. NADPH oxidase-derived superoxide anion mediates angiotensin II-enhanced carotid body chemoreceptor sensitivity in heart failure rabbits. Cardiovasc Res 2007;75:546-54.
- Takimoto E, Kass DA. Role of oxidative stress in cardiac hypertrophy and remodeling. Hypertension 2007;49:241-8.
- Yellon DM, Hausenloy DJ. Myocardial reperfusion injury. N Engl J Med 2007;357:1121-35.
- Perrelli MG, Pagliaro P, Penna C. Ischemia/reperfusion injury and cardioprotective mechanisms: Role of mitochondria and reactive oxygen species. World J Cardiol 2011;3:186-200.
- Jürgensen JS, Rosenberger C, Wiesener MS, et al. Persistent induction of HIF-1alpha and -2alpha in cardiomyocytes and stromal cells of ischemic myocardium. FASEB J 2004;18:1415-7.
- Jopling C, Suñè G, Faucherre A, et al. Hypoxia Induces Myocardial Regeneration in Zebrafish. Circulation 2012. [Epub ahead of print].
- Shiozawa Y, Havens AM, Pienta KJ, et al. The bone marrow niche: habitat to hematopoietic and mesenchymal stem cells, and unwitting host to molecular parasites. Leukemia 2008;22:941-50.
- Kavanagh DP, Kalia N. Hematopoietic stem cell homing to injured tissues. Stem Cell Rev 2011;7:672-82.
- Suárez-Álvarez B, Lopez-Vazquez A, Lopez-Larrea C. Mobilization and homing of hematopoietic stem cells. Adv Exp Med Biol 2012;741:152-70.
- Schulz C, von Andrian UH, Massberg S. Hematopoietic stem and progenitor cells: their mobilization and homing to bone marrow and peripheral tissue. Immunol Res 2009;44:160-8.
- Ghadge SK, Muhlstedt S, Ozcelik C, et al. SDF-1alpha as a therapeutic stem cell homing factor in myocardial infarction. Pharmacol Ther 2011;129:97-108.
- Smart N, Riley PR. The stem cell movement. Circ Res 2008;102:1155-68.
- Itou J, Oishi I, Kawakami H, et al. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development 2012;139:4133-42.
- Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Dev Biol 2009;336:137-44.
- Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol 2007;23:45-68.
- Lie-Venema H, van den Akker NM, Bax NA, et al. Origin, fate, and function of epicardium-derived cells (EPDCs) in normal and abnormal cardiac development. TheScientificWorldJournal 2007;7:1777-98.
- Ishii Y, Langberg J, Rosborough K, et al. Endothelial cell lineages of the heart. Cell Tissue Res 2009;335:67-73.
- Linask KK, Lash JW. Early heart development: dynamics of endocardial cell sorting suggests a common origin with cardiomyocytes. Dev Dyn 1993;196:62-9.
- Wu SM, Fujiwara Y, Cibulsky SM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell 2006;127:1137-50.
- Moretti A, Caron L, Nakano A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 2006;127:1151-65.
- Red-Horse K, Ueno H, Weissman IL, et al. Coronary arteries form by developmental reprogramming of venous cells. Nature 2010;464:549-53.
- Wu B, Zhang Z, Lui W, et al. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell 2012;151:1083-96.
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol 1996;174:221-32.
- Dettman RW, Denetclaw W Jr, Ordahl CP, et al. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol 1998;193:169-81.
- Männer J. Does the subepicardial mesenchyme contribute myocardioblasts to the myocardium of the chick embryo heart? A quail-chick chimera study tracing the fate of the epicardial primordium. Anat Rec 1999;255:212-26.
- Jiang X, Rowitch DH, Soriano P, et al. Fate of the mammalian cardiac neural crest. Development 2000;127:1607-16.
- Bersell K, Arab S, Haring B, et al. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 2009;138:257-70.
- Kühn B, del Monte F, Hajjar RJ, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med 2007;13:962-9.
- Schwartz SM, Haudenschild CC, Eddy EM. Endothelial regneration. I. Quantitative analysis of initial stages of endothelial regeneration in rat aortic intima. Lab Invest 1978;38:568-80.
- Fishman JA, Ryan GB, Karnovsky MJ. Endothelial regeneration in the rat carotid artery and the significance of endothelial denudation in the pathogenesis of myointimal thickening. Lab Invest 1975;32:339-51.
- Clopath P, Muller K, Staubli W, et al. In vivo and in vitro studies on endothelial regeneration. Haemostasis 1979;8:149-57.
- Itoh Y, Toriumi H, Yamada S, et al. Resident endothelial cells surrounding damaged arterial endothelium reendothelialize the lesion. Arterioscler Thromb Vasc Biol 2010;30:1725-32.
- Werner N, Priller J, Laufs U, et al. Bone marrow-derived progenitor cells modulate vascular reendothelialization and neointimal formation: effect of 3-hydroxy-3-methylglutaryl coenzyme a reductase inhibition. Arterioscler Thromb Vasc Biol 2002;22:1567-72.
- Werner N, Junk S, Laufs U, et al. Intravenous transfusion of endothelial progenitor cells reduces neointima formation after vascular injury. Circ Res 2003;93:e17-24.
- Hu Y, Zhang Z, Torsney E, et al. Abundant progenitor cells in the adventitia contribute to atherosclerosis of vein grafts in ApoE-deficient mice. J Clin Invest 2004;113:1258-65.
- Yoder MC. Is endothelium the origin of endothelial progenitor cells? Arterioscler Thromb Vasc Biol 2010;30:1094-103.
- Hagensen MK, Shim J, Falk E, et al. Flanking recipient vasculature, not circulating progenitor cells, contributes to endothelium and smooth muscle in murine allograft vasculopathy. Arterioscler Thromb Vasc Biol 2011;31:808-13.
- Hagensen MK, Shim J, Thim T, et al. Circulating endothelial progenitor cells do not contribute to plaque endothelium in murine atherosclerosis. Circulation 2010;121:898-905.
- Hagensen MK, Raarup MK, Mortensen MB, et al. Circulating endothelial progenitor cells do not contribute to regeneration of endothelium after murine arterial injury. Cardiovasc Res 2012;93:223-31.
- Limana F, Zacheo A, Mocini D, et al. Identification of myocardial and vascular precursor cells in human and mouse epicardium. Circ Res 2007;101:1255-65.
- Männer J. The development of pericardial villi in the chick embryo. Anat Embryol 1992;186:379-85.
- Männer J. Experimental study on the formation of the epicardium in chick embryos. Anat Embryol 1993;187:281-9.
- Loffredo FS, Steinhauser ML, Gannon J, et al. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell 2011;8:389-98.


