
Nomogram developed with selenoprotein S (SelS) genetic variation and clinical characteristics predicting risk of coronary artery disease in a Chinese population
Introduction
Coronary artery disease (CAD), caused by coronary artery stenosis or even occlusion, is the major cause of mortality and morbidity worldwide (1,2). CAD is a complex multifactorial disease and is independently associated with multiple risk factors such as advanced age, smoking, hyperlipidemia, diabetes, hypertension and chronic inflammation (3-7). Nevertheless, increasing evidences suggested that genetic factors play a vital role in the pathogenesis of CAD. Twin studies documented that about half of the overall risk to suffer from CAD are inherited. Furthermore, the most recent genome-wide association study (GWAS) study has increased the number of genome-wide significant single nucleotide polymorphisms (SNPs) for CAD to 58 (8).
Selenoprotein S (SelS) is a novel selenoprotein encoded by the SelS gene on chromosome 15q26.3 (9). SelS is located in both the plasma membranes and endoplasmic reticulum (ER) and especially expressed in adipose tissue, liver, skeletal muscle and heart (10,11). Recent researches have suggested that SelS may be involved in the development of diabetes, dyslipidemia and macrovascular complications (12,13). Walder et al. demonstrated that SelS was negatively correlated with blood glucose and serum insulin levels in a polygenic animal model of type 2 diabetes mellitus (T2DM)-Psammomys obesus (14). In addition, SelS has also been identified and shown to be functionally involved in the process of inflammatory response. Walder et al. firstly confirmed SelS as a receptor for the acute inflammatory response protein, serum amyloid A (SAA) and found that inhibition of SelS expression may upregulate the expression of SAA in HepG2 human liver cancer cells (14). All these suggested that SelS plays an important role in the inflammation and glycolipid metabolism, which both are risk factors for CAD.
Genetic factors may affect CAD through different mechanisms and individuals with specific genetic polymorphisms may be more susceptible to CAD. However, the association between SelS polymorphisms and CAD is unclear. In the present case-control study, we aimed to explore the association of SelS gene polymorphisms with CAD in a Chinese population. We present the following article in accordance with the Epidemiology (STROBE) Statement reporting checklist (available at http://dx.doi.org/10.21037/cdt-20-296).
Methods
Subjects
The study was approved by the Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University and informed consent was taken from all the patients (No. K201909-02). Five hundred and seventy-six subjects diagnosed with CAD were recruited at the First Affiliated Hospital of Xinjiang Medical University from January 2015 to December 2017. CAD was defined as the presence of stenosis of more than 50% luminal diameter in at least one significant coronary artery on coronary angiography. Subjects with spastic angina pectoris, infectious processes within 2 weeks, heart failure, adrenal dysfunction, and thyroid dysfunction were excluded. Four hundred and fifty-two subjects were recruited as control subjects and all control subjects were selected from volunteers who had angiographically normal coronary arteries and had no history of CAD. Coronary angiography in the control individuals was performed for the evaluation of chest pain. Subjects with CAD and any neoplasm, cardiomyopathy or severe illness limiting life expectance or refusing consent were excluded from control group. Hypertension was defined as a systolic blood pressure ≥140 mmHg and/or a diastolic blood pressure ≥90 mmHg at least on three distinct occasions. Subjects with T2DM were defined as those who had fasting plasma glucose (FPG) ≥7.0 mmol/L, or 2-h post-load plasma glucose (2hPG) ≥11.1 mmol/L, or had hypoglycemic therapy history. Smoking and drinking status was self-reported by study questionnaire.
Biochemical analysis
Blood samples were obtained from an antecubital vein into vacutainer tubes containing EDTA in the morning after an overnight fasting period. Serum concentrations of total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), triglycerides (TGs), apolipoprotein A1 (apoA1), lipoprotein A [Lp(a)] and glucose were measured using automated, clinically validated coulter.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using phenol and chloroform. Four SNPs (rs117613208, rs117512970, rs986500879 and rs542989868) were genotyped using direct sequencing method. The primers sequences were shown in Table 1. PCR amplification was performed using 10 µL of 2× Hieff PCR Master Mix, 1 µL of probes, 1 µL of DNA template and 8 µL ddH2O in a final reaction volume of 20 µL. Thermal cycling conditions were as follows: 95 °C for 5 min; 40 cycles of 94 °C for 30 s, 53 °C for 30 s, 72 °C for 40 s; and 72 °C for 5 min. Finally, 10% of the total genotyped samples were duplicated, and there was at least one positive and one negative control per 96-well DNA plate in our study. We obtained 100% concordance between the genotyped duplicate samples for each of the SNPs.

Full table
Development of GASDLY score
We utilized multivariable logistic regression analysis to identify the risk factors of CAD. SelS rs117613208 TT genotype, age, smoking, diabetes, hypertension, apoA1, and Lp(a) were identified independent risk factors. Therefore, we utilized these variables to develop a risk score named GASDLY score which was calculated as the following formula: GASDLY score =−2.145+ age ×0.59+ smoking ×1.675+ diabetes ×0.724+ SelS TT genotype ×0.745+ Lp(a) ×0.002−1.817× apoA1.
Statistical analysis
Analyses were carried out using SPSS version 22.0 (SPSS, Chicago, IL, USA). The Hardy-Weinberg equilibrium was assessed using χ2 analysis. Continuous variables were reported as mean ± SD and compared using Student’s t-test. Differences in enumeration data between CAD patients and control subjects were analyzed using χ2 analysis, as were differences in distributions of genotypes and alleles between CAD patients and control subjects. Logistic regression analyses with effect ratios [odds ratio (OR) and 95% confidence interval (CI)] were used to assess the contribution of the major risk factors. The receiver operating characteristic (ROC) curve was used to determine the sensitivity and specificity of GASDLY score for predicting CAD. Statistical significance was established at an alpha level of 0.05.
Results
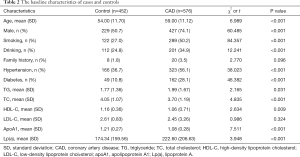
This study included 576 CAD cases (427 men, 149 women; mean age 59.00±11.12 years) and 452 controls (229 men, 223 women; mean age 54.00±11.70 years). The baseline characteristics of cases and controls are shown in Table 2. Age (P<0.001), male (P<0.001), smoking (P<0.001), drinking (P<0.001), hypertension (P<0.001), diabetes (P<0.001), TG (P=0.031), TC (P<0.001), HDL-C (P=0.009), apoA1 (P<0.001) and Lp(a) (P<0.001) were significantly different between CAD cases and controls.
Full table
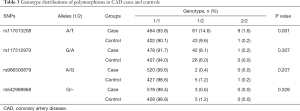
The genotype distributions of all polymorphisms in CAD cases and controls are shown in Table 3. The polymorphisms were all in Hardy-Weinberg equilibrium. The results show that the T allele of rs117613208 are more frequent in the CAD cases compared with that in the control group (P=0.001). However, there were no significant differences between CAD cases and controls in the distribution of rs117512970, rs986500879 and rs542989868 genotypes (P=0.307, P=0.207 and P=0.328, respectively).
Full table
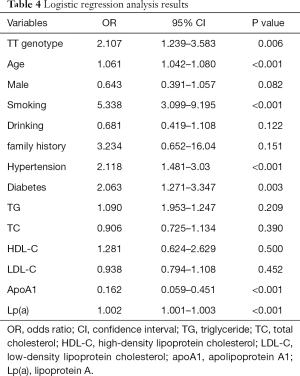
We further assessed the association between each SNP and CAD risk using a multivariable logistic regression model and rs117613208 was found to be an independent risk factor for CAD. Following the multivariate adjustments for the confounders, such as age, sex, smoking, drinking, family history, hypertension, diabetes, the plasma TG, TC, HDL-C, LDL-C, apoA1 and Lp(a) concentrations, the difference remained significant (P<0.006, OR =2.107, 95% CI: 1.239–3.583, Table 4).
Full table
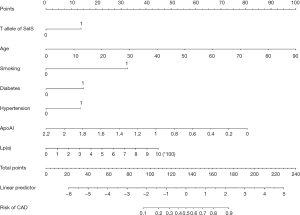
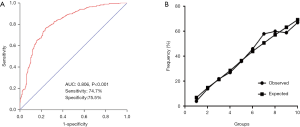
The nomogram was constructed using β-coefficients from the final Logistic multivariate model. The nomogram points for other variables were allocated according to the ratios of β-coefficients between rs117613208 and the selected variables (Figure 1). Furthermore, using SelS rs117613208, age, smoking, diabetes, hypertension, apoA1, and Lp(a) (GASDLY score), we developed a diagnostic model of CAD. As shown in Figure 2A, the AUC =0.806 (95% CI: 0.776–0.836, P<0.001), the sensitivity is 74.7% and the specificity is 75.5%). We also performed a calibration for the GASDLY score model using Hosmer-Lemeshow test (P=0.332). The results showed that the model prediction curve is very close to the actual observation curve, suggesting that the calibration ability of the model is excellent (Figure 2B).
Discussion
In the present study, we found that TT genotype of SelS was associated with CAD in a Chinese population. Furthermore, using clinical variables and TT genotype of SelS, we firstly developed a diagnostic model named GASDLY score which was of high discrimination and calibration to predict the risk of CAD.
Selenium (Se) is a trace element and an essential nutrient for human health (15). Despite the content of Se in human body is very low, Se is a key component of selenoproteins which is closely related to redox homeostasis, inflammatory regulation, myocardial and tumoral diseases. So far, 25 selenoproteins have been identified in the human proteome (16,17). SelS, as a member of the selenoprotein family, was first discovered in the liver of T2DM-Psammomys obesus (14). Recent researches have demonstrated that SelS gene polymorphisms were associated with serum levels of lipids, proinflammatory and anti-inflammatory cytokines, preeclampsia, rheumatoid arthritis, cancers and metabolic disorders (18-22). Although SelS have been identified as candidate genes leading to the risk of CAD, very few studies have examined its relationship to CAD in different populations. Alanne et al. performed two independent Finnish cohorts in 2,222 study subjects and found that the minor allele of rs8025174 in SelS gene were significantly associated with increased susceptibility to CAD in the combined analysis of the females in both cohorts. In addition, they also found that rs7178239 in SelS gene was associated with ischemic stroke in both sexes and both cohorts (19). Cox et al. performed a case-control study in 1,220 European American T2DM patients in the Diabetes Heart Disease Study to investigate the association between SelS genetic variants and subclinical cardiovascular disease (CVD) (21). Their result suggested that the SelS rs28665122, rs4965814, rs28628459, rs7178239 and rs12917258 in SelS gene were associated with subclinical CVD, while the SelS rs4965814, rs28628459 and rs9806366 were not.
In the current study, we performed a case-control study to evaluate the role of genetic variation of SelS on the risk of CAD in a Chinese Han population. The frequency of TT genotype in rs117613208 was significantly higher in the cases than that in the controls. Further multivariate analysis also showed that rs117613208was associated with increased susceptibility to CAD. However, we found no significant association between rs117512970, rs986500879 and rs542989868 and the risk of CAD in the present study. The mechanisms which may link the genetic variant of SelS to CAD are largely unclear. The possible pathophysiological rationales may be that SelS could increase CAD susceptibility by affecting the metabolism of glycolipid and inflammation. Walder et al. found that the expression of SelS is dysregulated in the liver of diabetic rats, suggesting the important role of SelS in T2DM (14,23). In vitro cultured HepG2 liver cancer cells, glucose may inhibit the expression of SelS and increase the expression of SelS may reduce the uptake of glucose. Gao et al. found that SelS is regulated by glucose deprivation and ER stress and demonstrated that glucose at concentrations lower than 5 nM may lead to the increasing expression of SelS in HepG2 cells (9). These results suggest that SelS plays an important role in the process of glucose metabolism. SelS was also confirmed as a receptor for the acute inflammatory response protein, SAA. Karlsson et al. found that SelS mRNA expression is positively associated with circulating SAA in human muscle and adipose tissue (24). Zeng et al. found that the expression of SAA in lipopolysaccharide (LPS)-stimulated HepG2 cells may upregulate when SelS was suppressed by small interfering RNA (siRNA) (25). In addition, inhibition of SelS expression could also upregulate the production and release of interleukin (IL)-1b, TNF-a and IL-6 in macrophages. Recent research performed by Ye found that inhibition of SelS gene expression siRNA may enhance ER stress and rendered vascular smooth muscle cells (VSMCs) more sensitive to hydrogen peroxide- or tunicamycin-induced injury and apoptosis, suggesting that SelS may affect atherosclerotic CVD by increasing the resistance of VSMCs to oxidative stress and ER stress (26). Nonetheless, there are some limitations to this study. The first limitation is the relatively sample size and a single-center experience study, the generalisability (external validity) of the study is still unclear. Second, the age and sex of the patients and the control group were not fully matched. Third, additional risk factors such as fibrinogen, plasma coagulation factors, or erythrocyte sedimentation rate were not evaluated to address the other confounding factors.
In conclusion, our study suggested the genetic polymorphism of SelS gene was associated with CAD. Furthermore, GASDLY score may be a novel diagnostic model for CAD in a Chinese population.
Acknowledgments
Funding: This research was funded by the National Natural Science Foundation of China (U1603381, 81760043), China Postdoctoral Science Foundation (2019 M652593), Xinjiang Science and Technology Aid Project (2019E0278), Tianshan Xuesong Project (2018XS17), and Henan Postdoctoral Science Foundation (1902006).
Footnote
Reporting Checklist: The authors present the study in accordance with the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/cdt-20-296
Data Sharing Statement: Available at http://dx.doi.org/10.21037/cdt-20-296
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cdt-20-296). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research was conducted in accordance with the Declaration of Helsinki. The study was approved by the Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University and informed consent was taken from all the patients (No. K201909-02).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Moran AE, Forouzanfar MH, Roth G, et al. The global burden of ischemic heart disease in 1990 and 2010: The Global Burden of Disease 2010 study. Circulation 2014;129:1493-501. [Crossref] [PubMed]
- Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015;386:743-800. [Crossref] [PubMed]
- Hammal F, Ezekowitz JA, Norris CM, et al. Smoking status and survival: impact on mortality of continuing to smoke one year after the angiographic diagnosis of coronary artery disease, a prospective cohort study. BMC Cardiovasc Disord 2014;14:133. [Crossref] [PubMed]
- Strissel KJ, Denis GV, Nikolajczyk BS. Immune regulators of inflammation in obesity-associated type 2 diabetes and coronary artery disease. Curr Opin Endocrinol Diabetes Obes 2014;21:330-8. [Crossref] [PubMed]
- Larsen SB, Grove EL, Wurtz M, et al. The influence of low-grade inflammation on platelets in patients with stable coronary artery disease. Thromb Haemost 2015;114:519-29. [Crossref] [PubMed]
- Aronson D, Edelman ER. Coronary artery disease and diabetes mellitus. Cardiol Clin 2014;32:439-55. [Crossref] [PubMed]
- Nakahara T, Dweck MR, Narula N, et al. Coronary artery calcification: from mechanism to molecular imaging. JACC Cardiovasc. Imaging 2017;10:582-593. [Crossref] [PubMed]
- Nikpay M, Goel A, Won HH, et al. A comprehensive 1000 genomes-based GWAS meta-analysis of coronary artery disease. Nat Genet 2015;47:1121-30. [Crossref] [PubMed]
- Gao Y, Walder K, Sunderland T, et al. Elevation in Tanis expression alters glucose metabolism and insulin sensitivity in H4IIE cells. Diabetes 2003;52:929-34. [Crossref] [PubMed]
- Speckmann B, Gerloff K, Simms L, et al. Selenoprotein S is a marker but not a regulator of endoplasmic reticulum stress in intestinal epithelial cells. Free Radic Biol Med 2014;67:265-77. [Crossref] [PubMed]
- Zhao Y, Li H, Men LL, et al. Effects of selenoprotein S on oxidative injury in human endothelial cells. J Transl Med 2013;11:287. [Crossref] [PubMed]
- Yu SS, Men LL, Wu JL, et al. The source of circulating selenoprotein S and its association with type 2 diabetes mellitus and atherosclerosis: a preliminary study. Cardiovasc Diabetol 2016;15:70. [Crossref] [PubMed]
- Qin HS, Yu PP, Sun Y, et al. Paclitaxel inhibits selenoprotein S expression and attenuates endoplasmic reticulum stress. Mol Med Rep 2016;13:5118-24. [Crossref] [PubMed]
- Walder K, Kantham L, McMillan JS, et al. Tanis: a link between type 2 diabetes and inflammation? Diabetes 2002;51:1859-66. [Crossref] [PubMed]
- Roman M, Jitaru P, Barbante C. Selenium biochemistry and its role for human health. Metallomics 2014;6:25-54. [Crossref] [PubMed]
- Behn D, Weiss-Nowak C, Kalcklösch M, et al. Studies on the distribution and characteristics of new mammalian selenium- containing proteins. Analyst 1995;120:823-5. [Crossref] [PubMed]
- Kryukov GV, Castellano S, Novoselov SV, et al. Characterization of mammalian selenoproteomes. Science 2003;300:1439-43. [Crossref] [PubMed]
- Curran JE, Jowett JB, Elliott KS, et al. Genetic variation in selenoprotein S influences inflammatory response. Nat Genet 2005;37:1234-41. [Crossref] [PubMed]
- Alanne M, Kristiansson K, Auro K. Variation in the selenoprotein S gene locus is associated with coronary heart disease and ischemic stroke in two independent Finnish cohorts. Hum Genet 2007;122:355-65. [Crossref] [PubMed]
- Olsson M, Olsson B, Jacobson P, et al. Expression of the selenoprotein S (SELS) gene in subcutaneous adipose tissue and SELS genotype are associated with metabolic risk factors. Metabolism 2011;60:114-20. [Crossref] [PubMed]
- Cox AJ, Lehtinen AB, Xu J, et al. Polymorphisms in the Selenoprotein S gene and subclinical cardiovascular disease in the Diabetes Heart Study. Acta Diabetol 2013;50:391-9. [Crossref] [PubMed]
- Gao Y, Xie X, Ma YT, et al. Genetic variation in Tanis was associated with elevating plasma triglyceride level in Chinese nondiabetic subjects. Lipids Health Dis 2013;12:97. [Crossref] [PubMed]
- Gao Y, Pagnon J, Feng HC, et al. Secretion of the glucose-regulated selenoprotein SEPS1 from hepatoma cells. Biochem Biophys Res Commun 2007;356:636-41. [Crossref] [PubMed]
- Karlsson HK, Tsuchida H, Lake S, et al. Relationship between serum amyloid A level and Tanis/SelS mRNA expression in skeletal muscle and adipose tissue from healthy and type 2 diabetic subjects. Diabetes 2004;53:1424-8. [Crossref] [PubMed]
- Zeng J, Du S, Zhou J, et al. Role of SelS in lipopolysaccharide-induced inflammatory response in hepatoma HepG2 cells. Arch Biochem Biophys 2008;478:1-6. [Crossref] [PubMed]
- Ye Y, Fu F, Li X, et al. Selenoprotein S Is Highly Expressed in the Blood Vessels and Prevents Vascular Smooth Muscle Cells from Apoptosis. J Cell Biochem 2016;117:106-17. [Crossref] [PubMed]


